Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing
- PMID: 23487774
- PMCID: PMC3607026
- DOI: 10.1073/pnas.1219247110
Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing
Abstract
The animal and human intestinal mucosa secretes an assortment of compounds to establish a physical barrier between the host tissue and intestinal contents, a separation that is vital for health. Some pathogenic microorganisms as well as members of the commensal intestinal microbiota have been shown to be able to break down these secreted compounds. Our understanding of host-compound degradation by the commensal microbiota has been limited to knowledge about simplified model systems because of the difficulty in studying the complex intestinal ecosystem in vivo. In this study, we introduce an approach that overcomes previous technical limitations and allows us to observe which microbial cells in the intestine use host-derived compounds. We added stable isotope-labeled threonine i.v. to mice and combined fluorescence in situ hybridization with high-resolution secondary ion mass spectrometry imaging to characterize utilization of host proteins by individual bacterial cells. We show that two bacterial species, Bacteroides acidifaciens and Akkermansia muciniphila, are important host-protein foragers in vivo. Using gnotobiotic mice we show that microbiota composition determines the magnitude and pattern of foraging by these organisms, demonstrating that a complex microbiota is necessary in order for this niche to be fully exploited. These results underscore the importance of in vivo studies of intestinal microbiota, and the approach presented in this study will be a powerful tool to address many other key questions in animal and human microbiome research.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
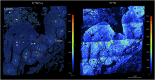

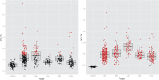
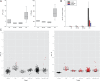
References
-
- Faure M, et al. Development of a rapid and convenient method to purify mucins and determine their in vivo synthesis rate in rats. Anal Biochem. 2002;307(2):244–251. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

