Dimerization of complement factor H-related proteins modulates complement activation in vivo
- PMID: 23487775
- PMCID: PMC3606973
- DOI: 10.1073/pnas.1219260110
Dimerization of complement factor H-related proteins modulates complement activation in vivo
Abstract
The complement system is a key component regulation influences susceptibility to age-related macular degeneration, meningitis, and kidney disease. Variation includes genomic rearrangements within the complement factor H-related (CFHR) locus. Elucidating the mechanism underlying these associations has been hindered by the lack of understanding of the biological role of CFHR proteins. Here we present unique structural data demonstrating that three of the CFHR proteins contain a shared dimerization motif and that this hitherto unrecognized structural property enables formation of both homodimers and heterodimers. Dimerization confers avidity for tissue-bound complement fragments and enables these proteins to efficiently compete with the physiological complement inhibitor, complement factor H (CFH), for ligand binding. Our data demonstrate that these CFHR proteins function as competitive antagonists of CFH to modulate complement activation in vivo and explain why variation in the CFHRs predisposes to disease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

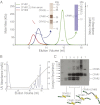
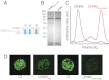
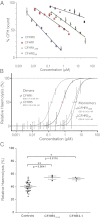
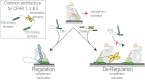
References
-
- de Cordoba SR, Tortajada A, Harris CL, Morgan BP. Complement dysregulation and disease: From genes and proteins to diagnostics and drugs. Immunobiology. 2012;217(11):1034–1046. - PubMed
-
- Abarrategui-Garrido C, Martínez-Barricarte R, López-Trascasa M, de Córdoba SR, Sánchez-Corral P. Characterization of complement factor H-related (CFHR) proteins in plasma reveals novel genetic variations of CFHR1 associated with atypical hemolytic uremic syndrome. Blood. 2009;114(19):4261–4271. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

