Chaperone activation by unfolding
- PMID: 23487787
- PMCID: PMC3619340
- DOI: 10.1073/pnas.1222458110
Chaperone activation by unfolding
Abstract
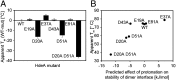
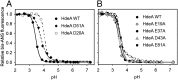
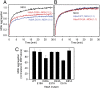
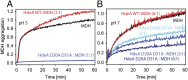
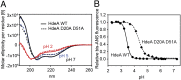
Conditionally disordered proteins can alternate between highly ordered and less ordered configurations under physiological conditions. Whereas protein function is often associated with the ordered conformation, for some of these conditionally unstructured proteins, the opposite applies: Their activation is associated with their unfolding. An example is the small periplasmic chaperone HdeA, which is critical for the ability of enteric bacterial pathogens like Escherichia coli to survive passage through extremely acidic environments, such as the human stomach. At neutral pH, HdeA is a chaperone-inactive dimer. On a shift to low pH, however, HdeA monomerizes, partially unfolds, and becomes rapidly active in preventing the aggregation of substrate proteins. By mutating two aspartic acid residues predicted to be responsible for the pH-dependent monomerization of HdeA, we have succeeded in isolating an HdeA mutant that is active at neutral pH. We find this HdeA mutant to be substantially destabilized, partially unfolded, and mainly monomeric at near-neutral pH at a concentration at which it prevents aggregation of a substrate protein. These results provide convincing evidence for direct activation of a protein by partial unfolding.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Comment in
-
How bacteria survive an acid trip.Proc Natl Acad Sci U S A. 2013 Apr 2;110(14):5279-80. doi: 10.1073/pnas.1303297110. Epub 2013 Mar 25. Proc Natl Acad Sci U S A. 2013. PMID: 23530238 Free PMC article. No abstract available.
References
-
- Smith JL. The role of gastric acid in preventing foodborne disease and how bacteria overcome acid conditions. J Food Prot. 2003;66(7):1292–1303. - PubMed
-
- Hong W, Wu YE, Fu X, Chang Z. Chaperone-dependent mechanisms for acid resistance in enteric bacteria. Trends Microbiol. 2012;20(7):328–335. - PubMed
-
- Zhao B, Houry WA. Acid stress response in enteropathogenic gammaproteobacteria: An aptitude for survival. Biochem Cell Biol. 2010;88(2):301–314. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

