Channel size conversion of Phi29 DNA-packaging nanomotor for discrimination of single- and double-stranded nucleic acids
- PMID: 23488809
- PMCID: PMC3663147
- DOI: 10.1021/nn400020z
Channel size conversion of Phi29 DNA-packaging nanomotor for discrimination of single- and double-stranded nucleic acids
Abstract
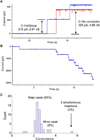
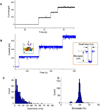
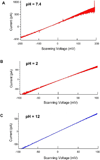
Nanopores have been utilized to detect the conformation and dynamics of polymers, including DNA and RNA. Biological pores are extremely reproducible at the atomic level with uniform channel sizes. The channel of the bacterial virus phi29 DNA-packaging motor is a natural conduit for the transportation of double-stranded DNA (dsDNA) and has the largest diameter among the well-studied biological channels. The larger channel facilitates translocation of dsDNA and offers more space for further channel modification and conjugation. Interestingly, the relatively large wild-type channel, which translocates dsDNA, cannot detect single-stranded nucleic acids (ssDNA or ssRNA) under the current experimental conditions. Herein, we reengineered this motor channel by removing the internal loop segment of the channel. The modification resulted in two classes of channels. One class was the same size as the wild-type channel, while the other class had a cross-sectional area about 60% of the wild-type. This smaller channel was able to detect the real-time translocation of single-stranded nucleic acids at single-molecule level. While the wild-type connector exhibited a one-way traffic property with respect to dsDNA translocation, the loop-deleted connector was able to translocate ssDNA and ssRNA with equal competencies from both termini. This finding of size alterations in reengineered motor channels expands the potential application of the phi29 DNA-packaging motor in nanomedicine, nanobiotechnology, and high-throughput single-pore DNA sequencing.
Figures
References
-
- Guo P. Structure and Function of Phi29 Hexameric RNA That Drive Viral DNA Packaging Motor: Review. Prog Nucleic Acid Res Mol Biol. 2002;72:415–472. - PubMed
-
- Simpson AA, Leiman PG, Tao Y, He Y, Badasso MO, Jardine PJ, Anderson DL, Rossman MG. Structure Determination of the Head-Tail Connector of Bacteriophage Phi29. Acta Cryst. 2001;D57:1260–1269. - PubMed
-
- Guasch A, Pous J, Ibarra B, Gomis-Ruth FX, Valpuesta JM, Sousa N, Carrascosa JL, Coll M. Detailed Architecture of a DNA Translocating Machine: the High-Resolution Structure of the Bacteriophage Phi29 Connector Particle. J. Mol. Biol. 2002;315:663–676. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources