The skin microbiome: current perspectives and future challenges
- PMID: 23489584
- PMCID: PMC3686918
- DOI: 10.1016/j.jaad.2013.01.016
The skin microbiome: current perspectives and future challenges
Abstract
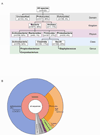
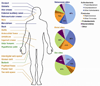
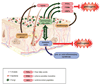
Complex communities of bacteria, fungi, and viruses thrive on our skin. The composition of these communities depends on skin characteristics, such as sebaceous gland concentration, moisture content, and temperature, as well as on host genetics and exogenous environmental factors. Recent metagenomic studies have uncovered a surprising diversity within these ecosystems and have fostered a new view of commensal organisms as playing a much larger role in immune modulation and epithelial health than previously expected. Understanding microbe-host interactions and discovering the factors that drive microbial colonization will help us understand the pathogenesis of skin diseases and develop new promicrobial and antimicrobial therapeutics.
Copyright © 2013 American Academy of Dermatology, Inc. Published by Mosby, Inc. All rights reserved.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures

References
-
- Holland KT, Cunliffe WJ, Roberts CD. Acne vulgaris: an investigation into the number of anaerobic diphtheroids and members of the Micrococcaceae in normal and acne skin. Br J Dermatol. 1977;96:623–626. - PubMed
-
- Thomsen RJ, Stranieri A, Knutson D, Strauss JS. Topical clindamycin treatment of acne. Clinical, surface lipid composition, and quantitative surface microbiology response. Arch Dermatol. 1980;116:1031–1034. - PubMed
-
- Till AE, Goulden V, Cunliffe WJ, Holland KT. The cutaneous microflora of adolescent, persistent and late-onset acne patients does not differ. Br J Dermatol. 2000;142:885–892. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

