A novel human endogenous retroviral protein inhibits cell-cell fusion
- PMID: 23492904
- PMCID: PMC3598002
- DOI: 10.1038/srep01462
A novel human endogenous retroviral protein inhibits cell-cell fusion
Abstract
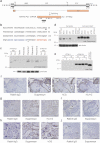
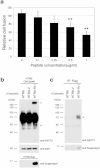
While common in viral infections and neoplasia, spontaneous cell-cell fusion, or syncytialization, is quite restricted in healthy tissues. Such fusion is essential to human placental development, where interactions between trophoblast-specific human endogenous retroviral (HERV) envelope proteins, called syncytins, and their widely-distributed cell surface receptors are centrally involved. We have identified the first host cell-encoded protein that inhibits cell fusion in mammals. Like the syncytins, this protein, called suppressyn, is HERV-derived, placenta-specific and well-conserved over simian evolution. In vitro, suppressyn binds to the syn1 receptor and inhibits syn1-, but not syn2-mediated trophoblast syncytialization. Suppressyn knock-down promotes cell-cell fusion in trophoblast cells and cell-associated and secreted suppressyn binds to the syn1 receptor, ASCT2. Identification of the first host cell-encoded inhibitor of mammalian cell fusion may encourage improved understanding of cell fusion mechanisms, of placental morphogenesis and of diseases resulting from abnormal cell fusion.
Figures


Similar articles
-
Could the Human Endogenous Retrovirus-Derived Syncytialization Inhibitor, Suppressyn, Limit Heterotypic Cell Fusion Events in the Decidua?Int J Mol Sci. 2021 Sep 23;22(19):10259. doi: 10.3390/ijms221910259. Int J Mol Sci. 2021. PMID: 34638599 Free PMC article.
-
Controlling Trophoblast Cell Fusion in the Human Placenta-Transcriptional Regulation of Suppressyn, an Endogenous Inhibitor of Syncytin-1.Biomolecules. 2023 Nov 7;13(11):1627. doi: 10.3390/biom13111627. Biomolecules. 2023. PMID: 38002309 Free PMC article.
-
Identification of the hASCT2-binding domain of the Env ERVWE1/syncytin-1 fusogenic glycoprotein.Retrovirology. 2006 Jul 4;3:41. doi: 10.1186/1742-4690-3-41. Retrovirology. 2006. PMID: 16820059 Free PMC article.
-
Syncytins expressed in human placental trophoblast.Placenta. 2021 Sep 15;113:8-14. doi: 10.1016/j.placenta.2021.01.006. Epub 2021 Jan 15. Placenta. 2021. PMID: 33504453 Free PMC article. Review.
-
Endogenous retroviral syncytin: compilation of experimental research on syncytin and its possible role in normal and disturbed human placentogenesis.Mol Hum Reprod. 2004 Aug;10(8):581-8. doi: 10.1093/molehr/gah070. Epub 2004 Jun 4. Mol Hum Reprod. 2004. PMID: 15181178 Review.
Cited by
-
gEVE: a genome-based endogenous viral element database provides comprehensive viral protein-coding sequences in mammalian genomes.Database (Oxford). 2016 May 30;2016:baw087. doi: 10.1093/database/baw087. Print 2016. Database (Oxford). 2016. PMID: 27242033 Free PMC article.
-
Cell Fusion and Syncytia Formation in Cancer.Results Probl Cell Differ. 2024;71:433-465. doi: 10.1007/978-3-031-37936-9_20. Results Probl Cell Differ. 2024. PMID: 37996689 Review.
-
Viral Causality of Human Cancer and Potential Roles of Human Endogenous Retroviruses in the Multi-Omics Era: An Evolutionary Epidemiology Review.Front Oncol. 2021 Oct 29;11:687631. doi: 10.3389/fonc.2021.687631. eCollection 2021. Front Oncol. 2021. PMID: 34778024 Free PMC article. Review.
-
Spatial multiomic landscape of the human placenta at molecular resolution.Nat Med. 2024 Dec;30(12):3495-3508. doi: 10.1038/s41591-024-03073-9. Epub 2024 Nov 20. Nat Med. 2024. PMID: 39567716
-
Receptor usage of Syncytin-1: ASCT2, but not ASCT1, is a functional receptor and effector of cell fusion in the human placenta.Proc Natl Acad Sci U S A. 2024 Oct 29;121(44):e2407519121. doi: 10.1073/pnas.2407519121. Epub 2024 Oct 21. Proc Natl Acad Sci U S A. 2024. PMID: 39432789 Free PMC article.
References
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

