Nearly finished genomes produced using gel microdroplet culturing reveal substantial intraspecies genomic diversity within the human microbiome
- PMID: 23493677
- PMCID: PMC3638143
- DOI: 10.1101/gr.142208.112
Nearly finished genomes produced using gel microdroplet culturing reveal substantial intraspecies genomic diversity within the human microbiome
Abstract
The majority of microbial genomic diversity remains unexplored. This is largely due to our inability to culture most microorganisms in isolation, which is a prerequisite for traditional genome sequencing. Single-cell sequencing has allowed researchers to circumvent this limitation. DNA is amplified directly from a single cell using the whole-genome amplification technique of multiple displacement amplification (MDA). However, MDA from a single chromosome copy suffers from amplification bias and a large loss of specificity from even very small amounts of DNA contamination, which makes assembling a genome difficult and completely finishing a genome impossible except in extraordinary circumstances. Gel microdrop cultivation allows culturing of a diverse microbial community and provides hundreds to thousands of genetically identical cells as input for an MDA reaction. We demonstrate the utility of this approach by comparing sequencing results of gel microdroplets and single cells following MDA. Bias is reduced in the MDA reaction and genome sequencing, and assembly is greatly improved when using gel microdroplets. We acquired multiple near-complete genomes for two bacterial species from human oral and stool microbiome samples. A significant amount of genome diversity, including single nucleotide polymorphisms and genome recombination, is discovered. Gel microdroplets offer a powerful and high-throughput technology for assembling whole genomes from complex samples and for probing the pan-genome of naturally occurring populations.
Figures
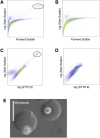

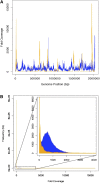
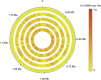
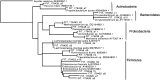
Comment in
-
Pathogen sequencing: Picking and choosing.Nat Rev Genet. 2013 May;14(5):304. doi: 10.1038/nrg3480. Epub 2013 Apr 9. Nat Rev Genet. 2013. PMID: 23568487 No abstract available.
-
Bacterial genomes by the droplet.Nat Methods. 2013 May;10(5):386. doi: 10.1038/nmeth.2469. Nat Methods. 2013. PMID: 23762908 No abstract available.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ 1990. Basic local alignment search tool. J Mol Biol 215: 403–410 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
