Molecular architecture and assembly of the eukaryotic proteasome
- PMID: 23495936
- PMCID: PMC3827779
- DOI: 10.1146/annurev-biochem-060410-150257
Molecular architecture and assembly of the eukaryotic proteasome
Abstract
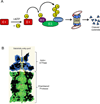
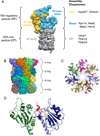
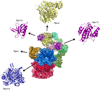
The eukaryotic ubiquitin-proteasome system is responsible for most aspects of regulatory and quality-control protein degradation in cells. Its substrates, which are usually modified by polymers of ubiquitin, are ultimately degraded by the 26S proteasome. This 2.6-MDa protein complex is separated into a barrel-shaped proteolytic 20S core particle (CP) of 28 subunits capped on one or both ends by a 19S regulatory particle (RP) comprising at least 19 subunits. The RP coordinates substrate recognition, removal of substrate polyubiquitin chains, and substrate unfolding and translocation into the CP for degradation. Although many atomic structures of the CP have been determined, the RP has resisted high-resolution analysis. Recently, however, a combination of cryo-electron microscopy, biochemical analysis, and crystal structure determination of several RP subunits has yielded a near-atomic-resolution view of much of the complex. Major new insights into chaperone-assisted proteasome assembly have also recently emerged. Here we review these novel findings.
Figures


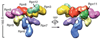


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

