Transcriptome exploration of the sex pheromone gland of Lutzomyia longipalpis (Diptera: Psychodidae: Phlebotominae)
- PMID: 23497448
- PMCID: PMC3632494
- DOI: 10.1186/1756-3305-6-56
Transcriptome exploration of the sex pheromone gland of Lutzomyia longipalpis (Diptera: Psychodidae: Phlebotominae)
Abstract
Background: Molecules involved in pheromone biosynthesis may represent alternative targets for insect population control. This may be particularly useful in managing the reproduction of Lutzomyia longipalpis, the main vector of the protozoan parasite Leishmania infantum in Latin America. Besides the chemical identity of the major components of the L. longipalpis sex pheromone, there is no information regarding the molecular biology behind its production. To understand this process, obtaining information on which genes are expressed in the pheromone gland is essential.
Methods: In this study we used a transcriptomic approach to explore the pheromone gland and adjacent abdominal tergites in order to obtain substantial general sequence information. We used a laboratory-reared L. longipalpis (one spot, 9-Methyl GermacreneB) population, captured in Lapinha Cave, state of Minas Gerais, Brazil for this analysis.
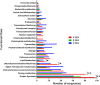
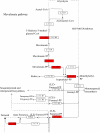
Results: From a total of 3,547 cDNA clones, 2,502 high quality sequences from the pheromone gland and adjacent tissues were obtained and assembled into 1,387 contigs. Through blast searches of public databases, a group of transcripts encoding proteins potentially involved in the production of terpenoid precursors were identified in the 4th abdominal tergite, the segment containing the pheromone gland. Among them, protein-coding transcripts for four enzymes of the mevalonate pathway such as 3-hydroxyl-3-methyl glutaryl CoA reductase, phosphomevalonate kinase, diphosphomevalonate descarboxylase, and isopentenyl pyrophosphate isomerase were identified. Moreover, transcripts coding for farnesyl diphosphate synthase and NADP+ dependent farnesol dehydrogenase were also found in the same tergite. Additionally, genes potentially involved in pheromone transportation were identified from the three abdominal tergites analyzed.
Conclusion: This study constitutes the first transcriptomic analysis exploring the repertoire of genes expressed in the tissue containing the L. longipalpis pheromone gland as well as the flanking tissues. Using a comparative approach, a set of molecules potentially present in the mevalonate pathway emerge as interesting subjects for further study regarding their association to pheromone biosynthesis. The sequences presented here may be used as a reference set for future research on pheromone production or other characteristics of pheromone communication in this insect. Moreover, some matches for transcripts of unknown function may provide fertile ground of an in-depth study of pheromone-gland specific molecules.
Figures






Similar articles
-
The Lutzomyia longipalpis complex: a brief natural history of aggregation-sex pheromone communication.Parasit Vectors. 2016 Nov 14;9(1):580. doi: 10.1186/s13071-016-1866-x. Parasit Vectors. 2016. PMID: 27842601 Free PMC article. Review.
-
Expression of the mevalonate pathway enzymes in the Lutzomyia longipalpis (Diptera: Psychodidae) sex pheromone gland demonstrated by an integrated proteomic approach.J Proteomics. 2014 Jan 16;96:117-32. doi: 10.1016/j.jprot.2013.10.028. Epub 2013 Nov 1. J Proteomics. 2014. PMID: 24185139 Free PMC article.
-
Pheromone gland development and pheromone production in lutzomyia longipalpis (Diptera: Psychodidae: Phlebotominae).J Med Entomol. 2011 May;48(3):489-95. doi: 10.1603/me10133. J Med Entomol. 2011. PMID: 21661306
-
Identification of the sex pheromone of Lutzomyia longipalpis (Lutz & Neiva, 1912) (Diptera: Psychodidae) from Asunción, Paraguay.Parasit Vectors. 2009 Nov 2;2(1):51. doi: 10.1186/1756-3305-2-51. Parasit Vectors. 2009. PMID: 19883505 Free PMC article.
-
Lutzomyia longipalpis (Diptera: Psychodidae: Phlebotominae): a review.An Acad Bras Cienc. 2003 Sep;75(3):301-30. doi: 10.1590/s0001-37652003000300005. Epub 2003 Aug 25. An Acad Bras Cienc. 2003. PMID: 12947480 Review.
Cited by
-
A diterpene synthase from the sandfly Lutzomyia longipalpis produces the pheromone sobralene.Proc Natl Acad Sci U S A. 2024 Mar 19;121(12):e2322453121. doi: 10.1073/pnas.2322453121. Epub 2024 Mar 12. Proc Natl Acad Sci U S A. 2024. PMID: 38470919 Free PMC article.
-
Novel family of terpene synthases evolved from trans-isoprenyl diphosphate synthases in a flea beetle.Proc Natl Acad Sci U S A. 2016 Mar 15;113(11):2922-7. doi: 10.1073/pnas.1523468113. Epub 2016 Mar 2. Proc Natl Acad Sci U S A. 2016. PMID: 26936952 Free PMC article.
-
The Lutzomyia longipalpis complex: a brief natural history of aggregation-sex pheromone communication.Parasit Vectors. 2016 Nov 14;9(1):580. doi: 10.1186/s13071-016-1866-x. Parasit Vectors. 2016. PMID: 27842601 Free PMC article. Review.
-
Differential gene expression underlying the biosynthesis of Dufour's gland signals in Bombus impatiens.Curr Res Insect Sci. 2023 Apr 5;3:100056. doi: 10.1016/j.cris.2023.100056. eCollection 2023. Curr Res Insect Sci. 2023. PMID: 37124651 Free PMC article.
-
Biological effects of trans, trans-farnesol in Leishmania amazonensis.Front Cell Infect Microbiol. 2023 Nov 16;13:1221246. doi: 10.3389/fcimb.2023.1221246. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 38035328 Free PMC article.
References
-
- Deane LM, Deane MP. Visceral leishmaniasis in Brazil: geographical distribution and transmission. Rev Inst Med Trop Sao Paulo. 1962;4:149–212. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

