Differential response of skeletal muscles to mTORC1 signaling during atrophy and hypertrophy
- PMID: 23497627
- PMCID: PMC3622636
- DOI: 10.1186/2044-5040-3-6
Differential response of skeletal muscles to mTORC1 signaling during atrophy and hypertrophy
Abstract
Background: Skeletal muscle mass is determined by the balance between protein synthesis and degradation. Mammalian target of rapamycin complex 1 (mTORC1) is a master regulator of protein translation and has been implicated in the control of muscle mass. Inactivation of mTORC1 by skeletal muscle-specific deletion of its obligatory component raptor results in smaller muscles and a lethal dystrophy. Moreover, raptor-deficient muscles are less oxidative through changes in the expression PGC-1α, a critical determinant of mitochondrial biogenesis. These results suggest that activation of mTORC1 might be beneficial to skeletal muscle by providing resistance to muscle atrophy and increasing oxidative function. Here, we tested this hypothesis by deletion of the mTORC1 inhibitor tuberous sclerosis complex (TSC) in muscle fibers.
Method: Skeletal muscles of mice with an acute or a permanent deletion of raptor or TSC1 were examined using histological, biochemical and molecular biological methods. Response of the muscles to changes in mechanical load and nerve input was investigated by ablation of synergistic muscles or by denervation .
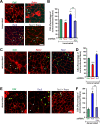
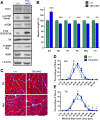
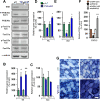
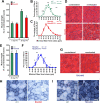
Results: Genetic deletion or knockdown of raptor, causing inactivation of mTORC1, was sufficient to prevent muscle growth and enhance muscle atrophy. Conversely, short-term activation of mTORC1 by knockdown of TSC induced muscle fiber hypertrophy and atrophy-resistance upon denervation, in both fast tibialis anterior (TA) and slow soleus muscles. Surprisingly, however, sustained activation of mTORC1 by genetic deletion of Tsc1 caused muscle atrophy in all but soleus muscles. In contrast, oxidative capacity was increased in all muscles examined. Consistently, TSC1-deficient soleus muscle was atrophy-resistant whereas TA underwent normal atrophy upon denervation. Moreover, upon overloading, plantaris muscle did not display enhanced hypertrophy compared to controls. Biochemical analysis indicated that the atrophy response of muscles was based on the suppressed phosphorylation of PKB/Akt via feedback inhibition by mTORC1 and subsequent increased expression of the E3 ubiquitin ligases MuRF1 and atrogin-1/MAFbx. In contrast, expression of both E3 ligases was not increased in soleus muscle suggesting the presence of compensatory mechanisms in this muscle.
Conclusions: Our study shows that the mTORC1- and the PKB/Akt-FoxO pathways are tightly interconnected and differentially regulated depending on the muscle type. These results indicate that long-term activation of the mTORC1 signaling axis is not a therapeutic option to promote muscle growth because of its strong feedback induction of the E3 ubiquitin ligases involved in protein degradation.
Figures


References
-
- Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3:1014–1019. doi: 10.1038/ncb1101-1014. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

