Dynamic integration of splicing within gene regulatory pathways
- PMID: 23498935
- PMCID: PMC3642998
- DOI: 10.1016/j.cell.2013.02.034
Dynamic integration of splicing within gene regulatory pathways
Abstract
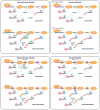
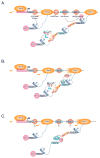
Precursor mRNA splicing is one of the most highly regulated processes in metazoan species. In addition to generating vast repertoires of RNAs and proteins, splicing has a profound impact on other gene regulatory layers, including mRNA transcription, turnover, transport, and translation. Conversely, factors regulating chromatin and transcription complexes impact the splicing process. This extensive crosstalk between gene regulatory layers takes advantage of dynamic spatial, physical, and temporal organizational properties of the cell nucleus, and further emphasizes the importance of developing a multidimensional understanding of splicing control.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures



References
-
- Ameur A, Zaghlool A, Halvardson J, Wetterbom A, Gyllensten U, Cavelier L, Feuk L. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat Struct Mol Biol. 2011;18:1435–1440. - PubMed
-
- Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau JC, et al. Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol. 2012;19:998–1004. - PubMed
-
- Amit M, Donyo M, Hollander D, Goren A, Kim E, Gelfman S, Lev-Maor G, Burstein D, Schwartz S, Postolsky B, et al. Differential GC content between exons and introns establishes distinct strategies of splice-site recognition. Cell Rep. 2012;1:543–556. - PubMed
-
- Auboeuf D, Honig A, Berget SM, O'Malley BW. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science. 2002;298:416–419. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

