Functional site plasticity in domain superfamilies
- PMID: 23499848
- PMCID: PMC3787744
- DOI: 10.1016/j.bbapap.2013.02.042
Functional site plasticity in domain superfamilies
Abstract
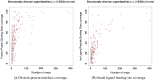
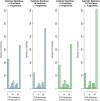
We present, to our knowledge, the first quantitative analysis of functional site diversity in homologous domain superfamilies. Different types of functional sites are considered separately. Our results show that most diverse superfamilies are very plastic in terms of the spatial location of their functional sites. This is especially true for protein-protein interfaces. In contrast, we confirm that catalytic sites typically occupy only a very small number of topological locations. Small-ligand binding sites are more diverse than expected, although in a more limited manner than protein-protein interfaces. In spite of the observed diversity, our results also confirm the previously reported preferential location of functional sites. We identify a subset of homologous domain superfamilies where diversity is particularly extreme, and discuss possible reasons for such plasticity, i.e. structural diversity. Our results do not contradict previous reports of preferential co-location of sites among homologues, but rather point at the importance of not ignoring other sites, especially in large and diverse superfamilies. Data on sites exploited by different relatives, within each well annotated domain superfamily, has been made accessible from the CATH website in order to highlight versatile superfamilies or superfamilies with highly preferential sites. This information is valuable for system biology and knowledge of any constraints on protein interactions could help in understanding the dynamic control of networks in which these proteins participate. The novelty of our work lies in the comprehensive nature of the analysis - we have used a significantly larger dataset than previous studies - and the fact that in many superfamilies we show that different parts of the domain surface are exploited by different relatives for ligand/protein interactions, particularly in superfamilies which are diverse in sequence and structure, an observation not previously reported on such a large scale. This article is part of a Special Issue entitled: The emerging dynamic view of proteins: Protein plasticity in allostery, evolution and self-assembly.
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures







References
-
- Ezkurdia L. Bartoli, Fariselli P., Casadio R., Valencia A., Tress M.L. Progress and challenges in predicting protein–protein interaction sites. Brief. Bioinform. 2009;10(3):233–246. - PubMed
-
- Aloy P., Ceulemans H., Stark A., Russell R.B. The relationship between sequence and interaction divergence in proteins. J. Mol. Biol. 2003;332:989–998. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

