The role of miRNAs in regulating gene expression networks
- PMID: 23500488
- PMCID: PMC3757117
- DOI: 10.1016/j.jmb.2013.03.007
The role of miRNAs in regulating gene expression networks
Abstract
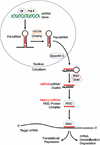
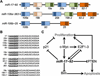
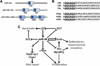
MicroRNAs (miRNAs) are key regulators of gene expression. They are conserved across species, expressed across cell types, and active against a large proportion of the transcriptome. The sequence-complementary mechanism of miRNA activity exploits combinatorial diversity, a property conducive to network-wide regulation of gene expression, and functional evidence supporting this hypothesized systems-level role has steadily begun to accumulate. The emerging models are exciting and will yield deep insight into the regulatory architecture of biology. However, because of the technical challenges facing the network-based study of miRNAs, many gaps remain. Here, we review mammalian miRNAs by describing recent advances in understanding their molecular activity and network-wide function.
Keywords: Ago2; Argonaute 2; Dicer; Drosha; EMT; ESC; KO; RISC; RNA-induced silencing complex; TGFβ; UTR; embryonic stem cell; epithelial-to-mesenchymal transition; knockout; let-7; miRNA; microRNA; network; pre-miRNA; precursor miRNA; transforming growth factor beta; untranslated region.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures



References
-
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. - PubMed
-
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. - PubMed
-
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

