"Candidatus Midichloriaceae" fam. nov. (Rickettsiales), an ecologically widespread clade of intracellular alphaproteobacteria
- PMID: 23503305
- PMCID: PMC3685259
- DOI: 10.1128/AEM.03971-12
"Candidatus Midichloriaceae" fam. nov. (Rickettsiales), an ecologically widespread clade of intracellular alphaproteobacteria
Abstract
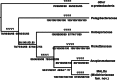
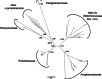
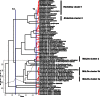
"Candidatus Midichloria mitochondrii" is an intramitochondrial bacterium of the order Rickettsiales associated with the sheep tick Ixodes ricinus. Bacteria phylogenetically related to "Ca. Midichloria mitochondrii" (midichloria and like organisms [MALOs]) have been shown to be associated with a wide range of hosts, from amoebae to a variety of animals, including humans. Despite numerous studies focused on specific members of the MALO group, no comprehensive phylogenetic and statistical analyses have so far been performed on the group as a whole. Here, we present a multidisciplinary investigation based on 16S rRNA gene sequences using both phylogenetic and statistical methods, thereby analyzing MALOs in the overall framework of the Rickettsiales. This study revealed that (i) MALOs form a monophyletic group; (ii) the MALO group is structured into distinct subgroups, verifying current genera as significant evolutionary units and identifying several subclades that could represent novel genera; (iii) the MALO group ranks at the level of described Rickettsiales families, leading to the proposal of the novel family "Candidatus Midichloriaceae." In addition, based on the phylogenetic trees generated, we present an evolutionary scenario to interpret the distribution and life history transitions of these microorganisms associated with highly divergent eukaryotic hosts: we suggest that aquatic/environmental protista have acted as evolutionary reservoirs for members of this novel family, from which one or more lineages with the capacity of infecting metazoa have evolved.
Figures


References
-
- Sassera D, Beninati T, Bandi C, Bouman EA, Sacchi L, Fabbi M, Lo N. 2006. ‘Candidatus Midichloria mitochondrii', an endosymbiont of the tick Ixodes ricinus with a unique intramitochondrial lifestyle. Int. J. Syst. Evol. Microbiol. 56:2535–2540 - PubMed
-
- Rizzoli A, Hauffe H, Carpi G, Vourc HG, Neteler M, Rosa R. 2011. Lyme borreliosis in Europe. Euro Surveill. 16:19906. - PubMed
-
- Dumler J, Walker D. 2005. Order II. Rickettsiales Gieszczykiewicz 1939, 25AL emend. Dumler, Barbet, Bekker, Dasch, Palmer, Ray, Rikihisa and Rurangirwa 2001, 2156, p 96–160. In Garrity GM, Brenner D, Krieg N, Staley J. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 2 Springer-Verlag, New York, NY
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

