Deciphering molecular circuits from genetic variation underlying transcriptional responsiveness to stimuli
- PMID: 23503680
- PMCID: PMC3622156
- DOI: 10.1038/nbt.2519
Deciphering molecular circuits from genetic variation underlying transcriptional responsiveness to stimuli
Abstract
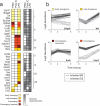
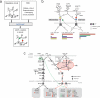
Individual genetic variation affects gene responsiveness to stimuli, often by influencing complex molecular circuits. Here we combine genomic and intermediate-scale transcriptional profiling with computational methods to identify variants that affect the responsiveness of genes to stimuli (responsiveness quantitative trait loci or reQTLs) and to position these variants in molecular circuit diagrams. We apply this approach to study variation in transcriptional responsiveness to pathogen components in dendritic cells from recombinant inbred mouse strains. We identify reQTLs that correlate with particular stimuli and position them in known pathways. For example, in response to a virus-like stimulus, a trans-acting variant responds as an activator of the antiviral response; using RNA interference, we identify Rgs16 as the likely causal gene. Our approach charts an experimental and analytic path to decipher the mechanisms underlying genetic variation in circuits that control responses to stimuli.
Figures
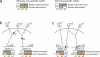



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

