myo-inositol and D-ribose ligand discrimination in an ABC periplasmic binding protein
- PMID: 23504019
- PMCID: PMC3650529
- DOI: 10.1128/JB.00116-13
myo-inositol and D-ribose ligand discrimination in an ABC periplasmic binding protein
Abstract
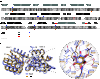
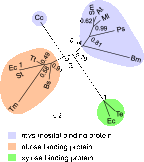
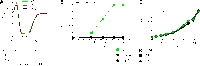
The periplasmic binding protein (PBP) IbpA mediates the uptake of myo-inositol by the IatP-IatA ATP-binding cassette transmembrane transporter. We report a crystal structure of Caulobacter crescentus IbpA bound to myo-inositol at 1.45 Å resolution. This constitutes the first structure of a PBP bound to inositol. IbpA adopts a type I PBP fold consisting of two α-β lobes that surround a central hinge. A pocket positioned between the lobes contains the myo-inositol ligand, which binds with submicromolar affinity (0.76 ± 0.08 μM). IbpA is homologous to ribose-binding proteins and binds D-ribose with low affinity (50.8 ± 3.4 μM). On the basis of IbpA and ribose-binding protein structures, we have designed variants of IbpA with inverted binding specificity for myo-inositol and D-ribose. Five mutations in the ligand-binding pocket are sufficient to increase the affinity of IbpA for D-ribose by 10-fold while completely abolishing binding to myo-inositol. Replacement of ibpA with these mutant alleles unable to bind myo-inositol abolishes C. crescentus growth in medium containing myo-inositol as the sole carbon source. Neither deletion of ibpA nor replacement of ibpA with the high-affinity ribose binding allele affected C. crescentus growth on D-ribose as a carbon source, providing evidence that the IatP-IatA transporter is specific for myo-inositol. This study outlines the evolutionary relationship between ribose- and inositol-binding proteins and provides insight into the molecular basis upon which these two related, but functionally distinct, classes of periplasmic proteins specifically bind carbohydrate ligands.
Figures




Similar articles
-
Genetic and computational identification of a conserved bacterial metabolic module.PLoS Genet. 2008 Dec;4(12):e1000310. doi: 10.1371/journal.pgen.1000310. Epub 2008 Dec 19. PLoS Genet. 2008. PMID: 19096521 Free PMC article.
-
Subcellular Localization Defects Characterize Ribose-Binding Mutant Proteins with New Ligand Properties in Escherichia coli.Appl Environ Microbiol. 2022 Jan 25;88(2):e0211721. doi: 10.1128/AEM.02117-21. Epub 2021 Nov 10. Appl Environ Microbiol. 2022. PMID: 34757821 Free PMC article.
-
In vitro reassembly of the ribose ATP-binding cassette transporter reveals a distinct set of transport complexes.J Biol Chem. 2015 Feb 27;290(9):5555-65. doi: 10.1074/jbc.M114.621573. Epub 2014 Dec 22. J Biol Chem. 2015. PMID: 25533465 Free PMC article.
-
HarmGR13 mediates myo-inositol taste perception in Helicoverpa armigera larvae.PLoS Genet. 2025 Jun 3;21(6):e1011744. doi: 10.1371/journal.pgen.1011744. eCollection 2025 Jun. PLoS Genet. 2025. PMID: 40460174 Free PMC article.
-
Potential role and therapeutic interests of myo-inositol in metabolic diseases.Biochimie. 2013 Oct;95(10):1811-27. doi: 10.1016/j.biochi.2013.05.011. Epub 2013 Jun 10. Biochimie. 2013. PMID: 23764390 Review.
Cited by
-
Native proteins trap high-energy transit conformations.Sci Adv. 2015 Oct 16;1(9):e1501188. doi: 10.1126/sciadv.1501188. eCollection 2015 Oct. Sci Adv. 2015. PMID: 26601321 Free PMC article.
-
Functional elucidation of TfuA in peptide backbone thioamidation.Nat Chem Biol. 2021 May;17(5):585-592. doi: 10.1038/s41589-021-00771-0. Epub 2021 Mar 11. Nat Chem Biol. 2021. PMID: 33707784 Free PMC article.
-
Current Blockades of Proteins inside Nanopores for Real-Time Metabolome Analysis.ACS Nano. 2020 Feb 25;14(2):2296-2307. doi: 10.1021/acsnano.9b09434. Epub 2020 Feb 7. ACS Nano. 2020. PMID: 32003969 Free PMC article.
-
Isolation and Characterization of Levoglucosan-Metabolizing Bacteria.Appl Environ Microbiol. 2022 Feb 22;88(4):e0186821. doi: 10.1128/AEM.01868-21. Epub 2021 Dec 15. Appl Environ Microbiol. 2022. PMID: 34910566 Free PMC article.
-
Conserved ABC Transport System Regulated by the General Stress Response Pathways of Alpha- and Gammaproteobacteria.J Bacteriol. 2017 Feb 14;199(5):e00746-16. doi: 10.1128/JB.00746-16. Print 2017 Mar 1. J Bacteriol. 2017. PMID: 27994018 Free PMC article.
References
-
- Nikaido H, Hall JA. 1998. Overview of bacterial ABC transporters. Methods Enzymol. 292:3–20 - PubMed
-
- Fischer M, Zhang QY, Hubbard RE, Thomas GH. 2010. Caught in a TRAP: substrate-binding proteins in secondary transport. Trends Microbiol. 18:471–478 - PubMed
-
- Rabus R, Jack DL, Kelly DJ, Saier MH., Jr 1999. TRAP transporters: an ancient family of extracytoplasmic solute-receptor-dependent secondary active transporters. Microbiology 145(Pt 12):3431–3445 - PubMed
-
- Winnen B, Hvorup RN, Saier MH., Jr 2003. The tripartite tricarboxylate transporter (TTT) family. Res. Microbiol. 154:457–465 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

