A stochastic simulator of birth-death master equations with application to phylodynamics
- PMID: 23505043
- PMCID: PMC3649681
- DOI: 10.1093/molbev/mst057
A stochastic simulator of birth-death master equations with application to phylodynamics
Abstract
In this article, we present a versatile new software tool for the simulation and analysis of stochastic models of population phylodynamics and chemical kinetics. Models are specified via an expressive and human-readable XML format and can be used as the basis for generating either single population histories or large ensembles of such histories. Importantly, phylogenetic trees or networks can be generated alongside the histories they correspond to, enabling investigations into the interplay between genealogies and population dynamics. Summary statistics such as means and variances can be recorded in place of the full ensemble, allowing for a reduction in the amount of memory used--an important consideration for models including large numbers of individual subpopulations or demes. In the case of population size histories, the resulting simulation output is written to disk in the flexible JSON format, which is easily read into numerical analysis environments such as R for visualization or further processing. Simulated phylogenetic trees can be recorded using the standard Newick or NEXUS formats, with extensions to these formats used for non-tree-like inheritance relationships.
Keywords: chemical kinetics simulation; epidemic modeling; phylogenetic trees; population genetics; stochastic simulation.
Figures


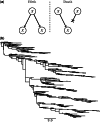
 , extending the age of the root substantially.
, extending the age of the root substantially.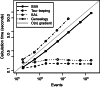
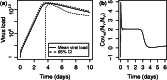
 (computations performed on an Intel Core i5 CPU operating at 3.0 GHz).
(computations performed on an Intel Core i5 CPU operating at 3.0 GHz).

References
-
- BEAST 2 Development Team. 2013. BEAST version 2 [Internet] Available from: http://beast2.cs.auckland.ac.nz (last accessed March 29, 2013)
-
- Britton T. Stochastic epidemic models: a survey. Math Biosci. 2010;225:24–35. - PubMed
-
- Couture-Beil A. 2013. rjson: JSON for R (version 0.2.12) [Internet] Available from: http://cran.r-project.org/web/packages/rjson/ (last accessed March 29, 2013)
-
- Drummond AJ, Pybus OG, Rambaut A, Forsberg R, Rodrigo AG. Measurably evolving populations. Trends Ecol Evol. 2003;18:481–488.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

