HAL: a hierarchical format for storing and analyzing multiple genome alignments
- PMID: 23505295
- PMCID: PMC3654707
- DOI: 10.1093/bioinformatics/btt128
HAL: a hierarchical format for storing and analyzing multiple genome alignments
Abstract
Motivation: Large multiple genome alignments and inferred ancestral genomes are ideal resources for comparative studies of molecular evolution, and advances in sequencing and computing technology are making them increasingly obtainable. These structures can provide a rich understanding of the genetic relationships between all subsets of species they contain. Current formats for storing genomic alignments, such as XMFA and MAF, are all indexed or ordered using a single reference genome, however, which limits the information that can be queried with respect to other species and clades. This loss of information grows with the number of species under comparison, as well as their phylogenetic distance.
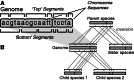
Results: We present HAL, a compressed, graph-based hierarchical alignment format for storing multiple genome alignments and ancestral reconstructions. HAL graphs are indexed on all genomes they contain. Furthermore, they are organized phylogenetically, which allows for modular and parallel access to arbitrary subclades without fragmentation because of rearrangements that have occurred in other lineages. HAL graphs can be created or read with a comprehensive C++ API. A set of tools is also provided to perform basic operations, such as importing and exporting data, identifying mutations and coordinate mapping (liftover).
Availability: All documentation and source code for the HAL API and tools are freely available at http://github.com/glennhickey/hal.
Contact: hickey@soe.ucsc.edu or haussler@soe.ucsc.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

