Distinct molecular strategies for Hox-mediated limb suppression in Drosophila: from cooperativity to dispensability/antagonism in TALE partnership
- PMID: 23505377
- PMCID: PMC3591290
- DOI: 10.1371/journal.pgen.1003307
Distinct molecular strategies for Hox-mediated limb suppression in Drosophila: from cooperativity to dispensability/antagonism in TALE partnership
Abstract
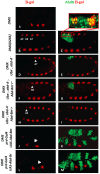
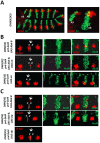
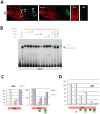
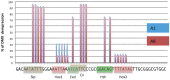
The emergence following gene duplication of a large repertoire of Hox paralogue proteins underlies the importance taken by Hox proteins in controlling animal body plans in development and evolution. Sequence divergence of paralogous proteins accounts for functional specialization, promoting axial morphological diversification in bilaterian animals. Yet functionally specialized paralogous Hox proteins also continue performing ancient common functions. In this study, we investigate how highly divergent Hox proteins perform an identical function. This was achieved by comparing in Drosophila the mode of limb suppression by the central (Ultrabithorax and AbdominalA) and posterior class (AbdominalB) Hox proteins. Results highlight that Hox-mediated limb suppression relies on distinct modes of DNA binding and a distinct use of TALE cofactors. Control of common functions by divergent Hox proteins, at least in the case studied, relies on evolving novel molecular properties. Thus, changes in protein sequences not only provide the driving force for functional specialization of Hox paralogue proteins, but also provide means to perform common ancient functions in distinct ways.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Pearson JC, Lemons D, McGinnis W (2005) Modulating Hox gene functions during animal body patterning. Nat Rev Genet 6: 893–904. - PubMed
-
- Merabet S, Hudry B, Saadaoui M, Graba Y (2009) Classification of sequence signatures: a guide to Hox protein function. Bioessays 31: 500–511. - PubMed
-
- Gehring WJ, Kloter U, Suga H (2009) Evolution of the Hox gene complex from an evolutionary ground state. Curr Top Dev Biol 88: 35–61. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

