FMR1 transcript isoforms: association with polyribosomes; regional and developmental expression in mouse brain
- PMID: 23505481
- PMCID: PMC3591412
- DOI: 10.1371/journal.pone.0058296
FMR1 transcript isoforms: association with polyribosomes; regional and developmental expression in mouse brain
Abstract
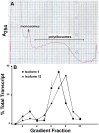
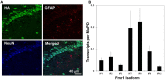
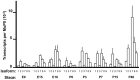
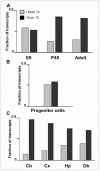
The primary transcript of the mammalian Fragile X Mental Retardation-1 gene (Fmr1), like many transcripts in the central nervous system, is alternatively spliced to yield mRNAs encoding multiple proteins, which can possess quite different biochemical properties. Despite the fact that the relative levels of the 12 Fmr1 transcript isoforms examined here vary by as much as two orders of magnitude amongst themselves in both adult and embryonic mouse brain, all are associated with polyribosomes, consistent with translation into the corresponding isoforms of the protein product, FMRP (Fragile X Mental Retardation Protein). Employing the RiboTag methodology developed in our laboratory, the relative proportions of the 7 most abundant transcript isoforms were measured specifically in neurons and found to be similar to those identified in whole brain. Measurements of isoform profiles across 11 regions of adult brain yielded similar distributions, with the exceptions of the hippocampus and the olfactory bulb. These two regions differ from most of the brain in relative amounts of transcripts encoding an alternate form of one of the KH RNA binding domains. A possible relationship between patterns of expression in the hippocampus and olfactory bulb and the presence of neuroblasts in these two regions is suggested by the isoform patterns in early embryonic brain and in cultured neural progenitor cells. These results demonstrate that the relative levels of the Fmr1 isoforms are modulated according to developmental stage, highlighting the complex ramifications of losing all the protein isoforms in individuals with Fragile X Syndrome. It should also be noted that, of the eight most prominent FMRP isoforms (1-3, 6-9 and 12) in mouse, only two have the major site of phosphorylation at Ser-499, which is thought to be involved in some of the regulatory interactions of this protein.
Conflict of interest statement
Figures



References
-
- Stetefeld J, Ruegg MA (2005) Structural and functional diversity generated by alternative mRNA splicing. Trends Biochem Sci 30: 515–521. - PubMed
-
- Lipscombe D (2005) Neuronal proteins custom designed by alternative splicing. Curr Opin Neurobiol 15: 358–363. - PubMed
-
- Raymond CK, Castle J, Garrett-Engele P, Armour CD, Kan Z, et al. (2004) Expression of alternatively spliced sodium channel alpha-subunit genes. Unique splicing patterns are observed in dorsal root ganglia. J Biol Chem 279: 46234–46241. - PubMed
-
- Ule J, Darnell RB (2006) RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr Opin Neurobiol 16: 102–110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

