Post-transcriptional regulation of connexin43 in H-Ras-transformed cells
- PMID: 23505521
- PMCID: PMC3594296
- DOI: 10.1371/journal.pone.0058500
Post-transcriptional regulation of connexin43 in H-Ras-transformed cells
Abstract
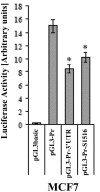
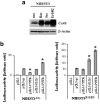
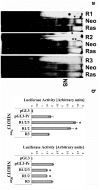
Connexin43 (Cx43) expression is lost in cancer cells and many studies have reported that Cx43 is a tumor suppressor gene. Paradoxically, in a cellular NIH3T3 model, we have previously shown that Ha-Ras-mediated oncogenic transformation results in increased Cx43 expression. Although the examination of transcriptional regulation revealed essential regulatory elements, it could not solve this paradox. Here we studied post-transcriptional regulation of Cx43 expression in cancer using the same model in search of novel gene regulatory elements. Upon Ras transformation, both Cx43 mRNA stability and translation efficiency were increased. We investigated the role of Cx43 mRNA 3' and 5'Untranslated regions (UTRs) and found an opposing effect; a 5'UTR-driven positive regulation is observed in Ras-transformed cells (NIH-3T3(Ras)), while the 3'UTR is active only in normal NIH-3T3(Neo) cells and completely silenced in NIH-3T3(Ras) cells. Most importantly, we identified a previously unknown regulatory element within the 3'UTR, named S1516, which accounts for this 3'UTR-mediated regulation. We also examined the effect of other oncogenes and found that Ras- and Src-transformed cells show a different Cx43 UTRs post-transcriptional regulation than ErbB2-transformed cells, suggesting distinct regulatory pathways. Next, we detected different patterns of S1516 RNA-protein complexes in NIH-3T3(Neo) compared to NIH-3T3(Ras) cells. A proteomic approach identified most of the S1516-binding proteins as factors involved in post-transcriptional regulation. Building on our new findings, we propose a model to explain the discrepancy between the Cx43 expression in Ras-transformed NIH3T3 cells and the data in clinical specimens.
Conflict of interest statement
Figures




Similar articles
-
Unexpected induction of the human connexin 43 promoter by the ras signaling pathway is mediated by a novel putative promoter sequence.Mol Pharmacol. 2003 Apr;63(4):821-31. doi: 10.1124/mol.63.4.821. Mol Pharmacol. 2003. PMID: 12644583
-
Downregulation of connexin43 by microRNA-130a in cardiomyocytes results in cardiac arrhythmias.J Mol Cell Cardiol. 2014 Sep;74:53-63. doi: 10.1016/j.yjmcc.2014.04.024. Epub 2014 May 10. J Mol Cell Cardiol. 2014. PMID: 24819345 Free PMC article.
-
Translational control of apolipoprotein B mRNA: regulation via cis elements in the 5' and 3' untranslated regions.Biochemistry. 2004 Jun 1;43(21):6734-44. doi: 10.1021/bi049887s. Biochemistry. 2004. PMID: 15157107
-
A position-specific 3'UTR sequence that accelerates mRNA decay.RNA Biol. 2016 Nov;13(11):1075-1077. doi: 10.1080/15476286.2016.1225645. Epub 2016 Aug 26. RNA Biol. 2016. PMID: 27565004 Free PMC article. Review.
-
The Untranslated Regions of mRNAs in Cancer.Trends Cancer. 2019 Apr;5(4):245-262. doi: 10.1016/j.trecan.2019.02.011. Epub 2019 Mar 22. Trends Cancer. 2019. PMID: 30961831 Free PMC article. Review.
Cited by
-
Connexin and pannexin channels in cancer.BMC Cell Biol. 2016 May 24;17 Suppl 1(Suppl 1):12. doi: 10.1186/s12860-016-0094-8. BMC Cell Biol. 2016. PMID: 27229305 Free PMC article. Review.
-
Specific functional pathologies of Cx43 mutations associated with oculodentodigital dysplasia.Mol Biol Cell. 2016 Jul 15;27(14):2172-85. doi: 10.1091/mbc.E16-01-0062. Epub 2016 May 25. Mol Biol Cell. 2016. PMID: 27226478 Free PMC article.
References
-
- Lundberg AS, Randell SH, Stewart SA, Elenbaas B, Hartwell KA, et al. (2002) Immortalization and transformation of primary human airway epithelial cells by gene transfer. Oncogene 21: 4577–4586. - PubMed
-
- Raponi M, Winkler H, Dracopoli NC (2008) KRAS mutations predict response to EGFR inhibitors. Curr Opin Pharmacol 8: 413–418. - PubMed
-
- Barbacid M (1987) ras genes. Annu Rev Biochem 56: 779–827. - PubMed
-
- Bos JL (1989) ras oncogenes in human cancer: a review. Cancer Res 49: 4682–4689. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

