Predicting where small molecules bind at protein-protein interfaces
- PMID: 23505538
- PMCID: PMC3591369
- DOI: 10.1371/journal.pone.0058583
Predicting where small molecules bind at protein-protein interfaces
Abstract
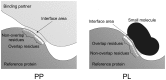
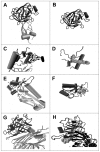
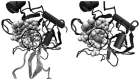
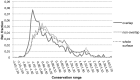
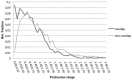
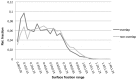
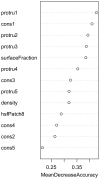
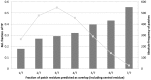
Small molecules that bind at protein-protein interfaces may either block or stabilize protein-protein interactions in cells. Thus, some of these binding interfaces may turn into prospective targets for drug design. Here, we collected 175 pairs of protein-protein (PP) complexes and protein-ligand (PL) complexes with known three-dimensional structures for which (1) one protein from the PP complex shares at least 40% sequence identity with the protein from the PL complex, and (2) the interface regions of these proteins overlap at least partially with each other. We found that those residues of the interfaces that may bind the other protein as well as the small molecule are evolutionary more conserved on average, have a higher tendency of being located in pockets and expose a smaller fraction of their surface area to the solvent than the remaining protein-protein interface region. Based on these findings we derived a statistical classifier that predicts patches at binding interfaces that have a higher tendency to bind small molecules. We applied this new prediction method to more than 10,000 interfaces from the protein data bank. For several complexes related to apoptosis the predicted binding patches were in direct contact to co-crystallized small molecules.
Conflict of interest statement
Figures




Similar articles
-
The distribution of ligand-binding pockets around protein-protein interfaces suggests a general mechanism for pocket formation.Proc Natl Acad Sci U S A. 2012 Mar 6;109(10):3784-9. doi: 10.1073/pnas.1117768109. Epub 2012 Feb 21. Proc Natl Acad Sci U S A. 2012. PMID: 22355140 Free PMC article.
-
Blind predictions of protein interfaces by docking calculations in CAPRI.Proteins. 2010 Nov 15;78(15):3085-95. doi: 10.1002/prot.22850. Proteins. 2010. PMID: 20839234
-
Protein-protein alternative binding modes do not overlap.Protein Sci. 2013 Aug;22(8):1141-5. doi: 10.1002/pro.2295. Epub 2013 Jul 3. Protein Sci. 2013. PMID: 23775945 Free PMC article.
-
Characterization and prediction of protein interfaces to infer protein-protein interaction networks.Curr Pharm Biotechnol. 2008 Apr;9(2):67-76. doi: 10.2174/138920108783955191. Curr Pharm Biotechnol. 2008. PMID: 18393863 Review.
-
Interaction-site prediction for protein complexes: a critical assessment.Bioinformatics. 2007 Sep 1;23(17):2203-9. doi: 10.1093/bioinformatics/btm323. Epub 2007 Jun 22. Bioinformatics. 2007. PMID: 17586545 Review.
Cited by
-
Small Molecule Antagonists of the DNA Repair ERCC1/XPA Protein-Protein Interaction.ChemMedChem. 2024 Apr 16;19(8):e202300648. doi: 10.1002/cmdc.202300648. Epub 2024 Mar 5. ChemMedChem. 2024. PMID: 38300970 Free PMC article.
-
Covalent Complex of DNA and Bacterial Topoisomerase: Implications in Antibacterial Drug Development.ChemMedChem. 2020 Apr 3;15(7):623-631. doi: 10.1002/cmdc.201900721. Epub 2020 Mar 18. ChemMedChem. 2020. PMID: 32043806 Free PMC article.
-
Small molecules, big targets: drug discovery faces the protein-protein interaction challenge.Nat Rev Drug Discov. 2016 Aug;15(8):533-50. doi: 10.1038/nrd.2016.29. Epub 2016 Apr 11. Nat Rev Drug Discov. 2016. PMID: 27050677 Review.
-
Composition of Overlapping Protein-Protein and Protein-Ligand Interfaces.PLoS One. 2015 Oct 30;10(10):e0140965. doi: 10.1371/journal.pone.0140965. eCollection 2015. PLoS One. 2015. PMID: 26517868 Free PMC article.
-
Considerations of Protein Subpockets in Fragment-Based Drug Design.Chem Biol Drug Des. 2016 Jan;87(1):5-20. doi: 10.1111/cbdd.12631. Epub 2015 Aug 31. Chem Biol Drug Des. 2016. PMID: 26307335 Free PMC article.
References
-
- Nooren IMA, Thornton JM (2003) Structural characterisation and functional significance of transient protein-protein interactions. J Mol Biol 325: 991–1018. - PubMed
-
- Yamada T, Bork P (2009) Evolution of biomolecular networks - lessons from metabolic and protein interactions. Nat Rev Mol Cell Biol 10: 791–803. - PubMed
-
- Klebe G (2009) Wirkstoffdesign: Entwurf und Wirkung von Arzneistoffen, Spektrum Akademischer Verlag 2nd ed.
-
- Wells JA, McClendon CL (2007) Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature 450: 1001–1009. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

