A comparative analysis of the properties of regulated promoter systems commonly used for recombinant gene expression in Escherichia coli
- PMID: 23506076
- PMCID: PMC3621392
- DOI: 10.1186/1475-2859-12-26
A comparative analysis of the properties of regulated promoter systems commonly used for recombinant gene expression in Escherichia coli
Abstract
Background: Production of recombinant proteins in bacteria for academic and commercial purposes is a well established field; however the outcomes of process developments for specific proteins are still often unpredictable. One reason is the limited understanding of the performance of expression cassettes relative to each other due to different genetic contexts. Here we report the results of a systematic study aiming at exclusively comparing commonly used regulator/promoter systems by standardizing the designs of the replicon backbones.
Results: The vectors used in this study are based on either the RK2- or the pMB1- origin of replication and contain the regulator/promoter regions of XylS/Pm (wild-type), XylS/Pm ML1-17 (a Pm variant), LacI/PT7lac, LacI/Ptrc and AraC/PBAD to control expression of different proteins with various origins. Generally and not unexpected high expression levels correlate with high replicon copy number and the LacI/PT7lac system generates more transcript than all the four other cassettes. However, this transcriptional feature does not always lead to a correspondingly more efficient protein production, particularly if protein functionality is considered. In most cases the XylS/Pm ML1-17 and LacI/PT7lac systems gave rise to the highest amounts of functional protein production, and the XylS/Pm ML1-17 is the most flexible in the sense that it does not require any specific features of the host. The AraC/PBAD system is very good with respect to tightness, and a commonly used bioinformatics prediction tool (RBS calculator) suggested that it has the most translation-efficient UTR. Expression was also studied by flow cytometry in individual cells, and the results indicate that cell to cell heterogeneity is very relevant for understanding protein production at the population level.
Conclusions: The choice of expression system needs to be evaluated for each specific case, but we believe that the standardized vectors developed for this study can be used to more easily identify the nature of case-specific bottlenecks. By then taking into account the relevant characteristics of each expression cassette it will be easier to make the best choice with respect to the goal of achieving high levels of protein expression in functional or non-functional form.
Figures

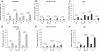

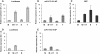


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

