1,2,3,4-Diepoxybutane-induced DNA-protein cross-linking in human fibrosarcoma (HT1080) cells
- PMID: 23506368
- PMCID: PMC3681427
- DOI: 10.1021/pr3011974
1,2,3,4-Diepoxybutane-induced DNA-protein cross-linking in human fibrosarcoma (HT1080) cells
Abstract
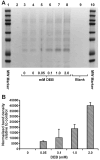
1,2,3,4-Diepoxybutane (DEB) is the key carcinogenic metabolite of 1,3-butadiene (BD), an important industrial and environmental chemical present in urban air and in cigarette smoke. DEB is a genotoxic bis-electrophile capable of cross-linking cellular biomolecules to form DNA-DNA and DNA-protein cross-links (DPCs). In the present work, mass spectrometry-based proteomics was employed to characterize DEB-mediated DNA-protein cross-linking in human fibrosarcoma (HT1080) cells. Over 150 proteins including histones, high mobility group proteins, transcription factors, splicing factors, and tubulins were found among those covalently cross-linked to chromosomal DNA in the presence of DEB. A large portion of the cross-linked proteins are known factors involved in DNA binding, transcriptional regulation, cell signaling, DNA repair, and DNA damage response. HPLC-ESI(+)-MS/MS analysis of total proteolytic digests revealed the presence of 1-(S-cysteinyl)-4-(guan-7-yl)-2,3-butanediol conjugates, confirming that DEB forms DPCs between cysteine thiols within proteins and the N-7 guanine positions within DNA. However, relatively high concentrations of DEB were required to achieve significant DPC formation, indicating that it is a poor cross-linking agent as compared to antitumor nitrogen mustards and platinum compounds.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

