Gene expression profiling of breast cancer survivability by pooled cDNA microarray analysis using logistic regression, artificial neural networks and decision trees
- PMID: 23506640
- PMCID: PMC3614553
- DOI: 10.1186/1471-2105-14-100
Gene expression profiling of breast cancer survivability by pooled cDNA microarray analysis using logistic regression, artificial neural networks and decision trees
Abstract
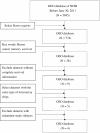
Background: Microarray technology can acquire information about thousands of genes simultaneously. We analyzed published breast cancer microarray databases to predict five-year recurrence and compared the performance of three data mining algorithms of artificial neural networks (ANN), decision trees (DT) and logistic regression (LR) and two composite models of DT-ANN and DT-LR. The collection of microarray datasets from the Gene Expression Omnibus, four breast cancer datasets were pooled for predicting five-year breast cancer relapse. After data compilation, 757 subjects, 5 clinical variables and 13,452 genetic variables were aggregated. The bootstrap method, Mann-Whitney U test and 20-fold cross-validation were performed to investigate candidate genes with 100 most-significant p-values. The predictive powers of DT, LR and ANN models were assessed using accuracy and the area under ROC curve. The associated genes were evaluated using Cox regression.
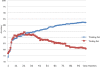
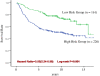
Results: The DT models exhibited the lowest predictive power and the poorest extrapolation when applied to the test samples. The ANN models displayed the best predictive power and showed the best extrapolation. The 21 most-associated genes, as determined by integration of each model, were analyzed using Cox regression with a 3.53-fold (95% CI: 2.24-5.58) increased risk of breast cancer five-year recurrence.
Conclusions: The 21 selected genes can predict breast cancer recurrence. Among these genes, CCNB1, PLK1 and TOP2A are in the cell cycle G2/M DNA damage checkpoint pathway. Oncologists can offer the genetic information for patients when understanding the gene expression profiles on breast cancer recurrence.
Figures

Similar articles
-
An Efficient Mixed-Model for Screening Differentially Expressed Genes of Breast Cancer Based on LR-RF.IEEE/ACM Trans Comput Biol Bioinform. 2019 Jan-Feb;16(1):124-130. doi: 10.1109/TCBB.2018.2829519. Epub 2018 Apr 23. IEEE/ACM Trans Comput Biol Bioinform. 2019. PMID: 29993693
-
Statistical characterization and classification of colon microarray gene expression data using multiple machine learning paradigms.Comput Methods Programs Biomed. 2019 Jul;176:173-193. doi: 10.1016/j.cmpb.2019.04.008. Epub 2019 Apr 10. Comput Methods Programs Biomed. 2019. PMID: 31200905
-
Comparison of three data mining models for prediction of advanced schistosomiasis prognosis in the Hubei province.PLoS Negl Trop Dis. 2018 Feb 15;12(2):e0006262. doi: 10.1371/journal.pntd.0006262. eCollection 2018 Feb. PLoS Negl Trop Dis. 2018. PMID: 29447165 Free PMC article.
-
Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care?Nat Rev Cancer. 2007 Jul;7(7):545-53. doi: 10.1038/nrc2173. Nat Rev Cancer. 2007. PMID: 17585334 Review.
-
[Approach to the methodology of classification and regression trees].Gac Sanit. 2008 Jan-Feb;22(1):65-72. doi: 10.1157/13115113. Gac Sanit. 2008. PMID: 18261446 Review. Spanish.
Cited by
-
A comparative evaluation of data-merging and meta-analysis methods for reconstructing gene-gene interactions.BMC Bioinformatics. 2016 Jun 6;17 Suppl 5(Suppl 5):194. doi: 10.1186/s12859-016-1038-1. BMC Bioinformatics. 2016. PMID: 27294826 Free PMC article.
-
The effect of centromere protein U silencing by lentiviral mediated RNA interference on the proliferation and apoptosis of breast cancer.Oncol Lett. 2018 Nov;16(5):6721-6728. doi: 10.3892/ol.2018.9477. Epub 2018 Sep 21. Oncol Lett. 2018. PMID: 30405814 Free PMC article.
-
Genetic co-expression networks contribute to creating predictive model and exploring novel biomarkers for the prognosis of breast cancer.Sci Rep. 2021 Mar 31;11(1):7268. doi: 10.1038/s41598-021-84995-z. Sci Rep. 2021. PMID: 33790307 Free PMC article.
-
Cascaded Wx: A Novel Prognosis-Related Feature Selection Framework in Human Lung Adenocarcinoma Transcriptomes.Front Genet. 2019 Jul 19;10:662. doi: 10.3389/fgene.2019.00662. eCollection 2019. Front Genet. 2019. PMID: 31379926 Free PMC article.
-
BAG3 Overexpression and Cytoprotective Autophagy Mediate Apoptosis Resistance in Chemoresistant Breast Cancer Cells.Neoplasia. 2018 Mar;20(3):263-279. doi: 10.1016/j.neo.2018.01.001. Epub 2018 Feb 22. Neoplasia. 2018. PMID: 29462756 Free PMC article.
References
-
- American Cancer Society. 2011. http://www.cancer.org/docroot/home/index.asp.
-
- Eifel P, Axelson JA, Costa J, Crowley J, Curran WJ Jr, Deshler A, Fulton S, Hendricks CB, Kemeny M, Kornblith AB. National Institutes of Health Consensus Development Conference Statement: adjuvant therapy for breast cancer: 1–3 November 2000. J Natl Cancer Inst. 2001;93:979–989. - PubMed
-
- Gemignani F, Perra C, Landi S, Canzian F, Kurg A, Tonisson N, Galanello R, Cao A, Metspalu A, Romeo G. Reliable detection of beta-thalassemia and G6PD mutations by a DNA microarray. Clin Chem. 2002;48:2051–2054. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

