Target identification and mechanism of action in chemical biology and drug discovery
- PMID: 23508189
- PMCID: PMC5543995
- DOI: 10.1038/nchembio.1199
Target identification and mechanism of action in chemical biology and drug discovery
Abstract
Target-identification and mechanism-of-action studies have important roles in small-molecule probe and drug discovery. Biological and technological advances have resulted in the increasing use of cell-based assays to discover new biologically active small molecules. Such studies allow small-molecule action to be tested in a more disease-relevant setting at the outset, but they require follow-up studies to determine the precise protein target or targets responsible for the observed phenotype. Target identification can be approached by direct biochemical methods, genetic interactions or computational inference. In many cases, however, combinations of approaches may be required to fully characterize on-target and off-target effects and to understand mechanisms of small-molecule action.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


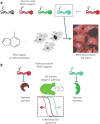


References
-
- Sundberg SA. High-throughput and ultra-high-throughput screening: solution- and cell-based approaches. Curr Opin Biotechnol. 2000;11:47–53. - PubMed
-
- Mayr LM, Bojanic D. Novel trends in high-throughput screening. Curr Opin Pharmacol. 2009;9:580–588. - PubMed
-
- Koehn FE. High impact technologies for natural products screening. Prog Drug Res. 2008;65(175):177–210. - PubMed
-
- Lachance H, Wetzel S, Kumar K, Waldmann H. Charting, navigating, and populating natural product chemical space for drug discovery. J Med Chem. 2012;55:5989–6001. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

