Modulation of BK channel by MicroRNA-9 in neurons after exposure to HIV and methamphetamine
- PMID: 23508624
- PMCID: PMC3715589
- DOI: 10.1007/s11481-013-9446-8
Modulation of BK channel by MicroRNA-9 in neurons after exposure to HIV and methamphetamine
Abstract
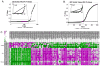
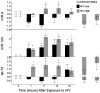
MicroRNAs (miR) regulate phenotype and function of neurons by binding to miR-response elements (MRE) in the 3' untranslated regions (3'UTR) of various messenger RNAs to inhibit translation. MiR expression can be induced or inhibited by environmental factors like drug exposure and viral infection, leading to changes in cellular physiology. We hypothesized that the effects of methamphetamine (MA) and human immunodeficiency virus (HIV)-infection in the brain will induce changes in miR expression, and have downstream regulatory consequences in neurons. We first used a PCR-based array to screen for differential expression of 380 miRs in frontal cortex autopsy tissues of HIV-positive MA abusers and matched controls. These results showed significantly increased expression of the neuron-specific miR-9. In vitro, we used SH-SY5Y cells, an experimental system for dopaminergic studies, to determine miR expression by quantitative PCR after exposure to MA in the presence or absence of conditioned media from HIV-infected macrophages. Again, we found that miR-9 was significantly increased compared to controls. We also examined the inwardly rectifying potassium channel, KCNMA1, which has alternative splice variants that contain an MRE to miR-9. We identified alternate 3'UTRs of KCNMA1 both in vitro and in the autopsy specimens and found differential splice variant expression of KCNMA1, operating via the increased miR-9. Our results suggest that HIV and MA -induced elevated miR-9, leading to suppression of MRE-containing splice variants of KCNMA1, which may affect neurotransmitter release in dopaminergic neurons.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
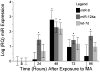



References
-
- Agrawal L, Louboutin JP, Marusich E, Reyes BA, Van Bockstaele EJ, Strayer DS. Dopaminergic neurotoxicity of HIV-1 gp120: reactive oxygen species as signaling intermediates. Brain Res. 2010;1306:116–130. - PubMed
-
- Aylward EH, Henderer JD, McArthur JC, Brettschneider PD, Harris GJ, Barta PE, Pearlson GD. Reduced basal ganglia volume in HIV-1-associated dementia: results from quantitative neuroimaging. Neurology. 1993;43:2099–2104. - PubMed
-
- Banerjee S, Neveu P, Kosik KS. A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron. 2009;64:871–884. - PubMed
-
- Basso MR, Bornstein RA. Neurobehavioural consequences of substance abuse and HIV infection. J Psychopharmacol. 2000;14:228–237. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

