Pericentric chromatin loops function as a nonlinear spring in mitotic force balance
- PMID: 23509068
- PMCID: PMC3601350
- DOI: 10.1083/jcb.201208163
Pericentric chromatin loops function as a nonlinear spring in mitotic force balance
Abstract
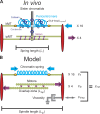
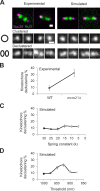
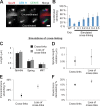
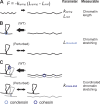
The mechanisms by which sister chromatids maintain biorientation on the metaphase spindle are critical to the fidelity of chromosome segregation. Active force interplay exists between predominantly extensional microtubule-based spindle forces and restoring forces from chromatin. These forces regulate tension at the kinetochore that silences the spindle assembly checkpoint to ensure faithful chromosome segregation. Depletion of pericentric cohesin or condensin has been shown to increase the mean and variance of spindle length, which have been attributed to a softening of the linear chromatin spring. Models of the spindle apparatus with linear chromatin springs that match spindle dynamics fail to predict the behavior of pericentromeric chromatin in wild-type and mutant spindles. We demonstrate that a nonlinear spring with a threshold extension to switch between spring states predicts asymmetric chromatin stretching observed in vivo. The addition of cross-links between adjacent springs recapitulates coordination between pericentromeres of neighboring chromosomes.
Figures


References
-
- Batchelor G.K. 1967. An Introduction to Fluid Dynamics. Cambridge University Press, Cambridge, UK: 615 pp
-
- Birtwistle M.R., Rauch J., Kiyatkin A., Aksamitiene E., Dobrzyński M., Hoek J.B., Kolch W., Ogunnaike B.A., Kholodenko B.N. 2012. Emergence of bimodal cell population responses from the interplay between analog single-cell signaling and protein expression noise. BMC Syst. Biol. 6:109 10.1186/1752-0509-6-109 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

