A novel single-cell screening platform reveals proteome plasticity during yeast stress responses
- PMID: 23509072
- PMCID: PMC3601363
- DOI: 10.1083/jcb.201301120
A novel single-cell screening platform reveals proteome plasticity during yeast stress responses
Erratum in
- J Cell Biol. 2013 Apr 15;201(2):353
Abstract
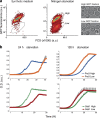
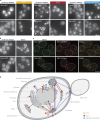
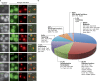
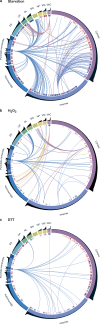
Uncovering the mechanisms underlying robust responses of cells to stress is crucial for our understanding of cellular physiology. Indeed, vast amounts of data have been collected on transcriptional responses in Saccharomyces cerevisiae. However, only a handful of pioneering studies describe the dynamics of proteins in response to external stimuli, despite the fact that regulation of protein levels and localization is an essential part of such responses. Here we characterized unprecedented proteome plasticity by systematically tracking the localization and abundance of 5,330 yeast proteins at single-cell resolution under three different stress conditions (DTT, H2O2, and nitrogen starvation) using the GFP-tagged yeast library. We uncovered a unique "fingerprint" of changes for each stress and elucidated a new response arsenal for adapting to radical environments. These include bet-hedging strategies, organelle rearrangement, and redistribution of protein localizations. All data are available for download through our online database, LOQATE (localization and quantitation atlas of yeast proteome).
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

