Additional pathway to translate the downstream ndhK cistron in partially overlapping ndhC-ndhK mRNAs in chloroplasts
- PMID: 23509265
- PMCID: PMC3619338
- DOI: 10.1073/pnas.1219914110
Additional pathway to translate the downstream ndhK cistron in partially overlapping ndhC-ndhK mRNAs in chloroplasts
Abstract
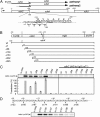
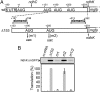
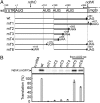
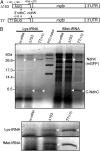
The chloroplast NAD(P)H dehydrogenase (NDH) C (ndhC) and ndhK genes partially overlap and are cotranscribed in many plants. We previously reported that the tobacco ndhC/K genes are translationally coupled but produce NdhC and NdhK, subunits of the NDH complex, in similar amounts. Generally, translation of the downstream cistron in overlapping mRNAs is very low. Hence, these findings suggested that the ndhK cistron is translated not only from the ndhC 5'UTR but also by an additional pathway. Using an in vitro translation system from tobacco chloroplasts, we report here that free ribosomes enter, with formylmethionyl-tRNA(fMet), at an internal AUG start codon that is located in frame in the middle of the upstream ndhC cistron, translate the 3' half of the ndhC cistron, reach the ndhK start codon, and that, at that point, some ribosomes resume ndhK translation. We detected a peptide corresponding to a 57-amino-acid product encoded by the sequence from the internal AUG to the ndhC stop codon. We propose a model in which the internal initiation site AUG is not designed for synthesizing a functional isoform but for delivering additional ribosomes to the ndhK cistron to produce NdhK in the amount required for the assembly of the NDH complex. This pathway is a unique type of translation to produce protein in the needed amount with the cost of peptide synthesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Termination codon-dependent translation of partially overlapping ndhC-ndhK transcripts in chloroplasts.Proc Natl Acad Sci U S A. 2008 Dec 9;105(49):19550-4. doi: 10.1073/pnas.0809240105. Epub 2008 Nov 25. Proc Natl Acad Sci U S A. 2008. PMID: 19033452 Free PMC article.
-
Translation of partially overlapping psbD-psbC mRNAs in chloroplasts: the role of 5'-processing and translational coupling.Nucleic Acids Res. 2012 Apr;40(7):3152-8. doi: 10.1093/nar/gkr1185. Epub 2011 Dec 8. Nucleic Acids Res. 2012. PMID: 22156163 Free PMC article.
-
The downstream atpE cistron is efficiently translated via its own cis-element in partially overlapping atpB-atpE dicistronic mRNAs in chloroplasts.Nucleic Acids Res. 2011 Nov;39(21):9405-12. doi: 10.1093/nar/gkr644. Epub 2011 Aug 16. Nucleic Acids Res. 2011. PMID: 21846772 Free PMC article.
-
Plastid mRNA translation.Methods Mol Biol. 2014;1132:73-91. doi: 10.1007/978-1-62703-995-6_4. Methods Mol Biol. 2014. PMID: 24599847 Review.
-
Pushing the limits of the scanning mechanism for initiation of translation.Gene. 2002 Oct 16;299(1-2):1-34. doi: 10.1016/s0378-1119(02)01056-9. Gene. 2002. PMID: 12459250 Free PMC article. Review.
Cited by
-
Seamless editing of the chloroplast genome in plants.BMC Plant Biol. 2016 Jul 29;16(1):168. doi: 10.1186/s12870-016-0857-6. BMC Plant Biol. 2016. PMID: 27474038 Free PMC article.
-
Full transcription of the chloroplast genome in photosynthetic eukaryotes.Sci Rep. 2016 Jul 26;6:30135. doi: 10.1038/srep30135. Sci Rep. 2016. PMID: 27456469 Free PMC article.
-
IAOseq: inferring abundance of overlapping genes using RNA-seq data.BMC Bioinformatics. 2015;16 Suppl 1(Suppl 1):S3. doi: 10.1186/1471-2105-16-S1-S3. Epub 2015 Jan 21. BMC Bioinformatics. 2015. PMID: 25707673 Free PMC article.
References
-
- Sugiura M. The chloroplast genome. Plant Mol Biol. 1992;19(1):149–168. - PubMed
-
- Yukawa M, Tsudzuki T, Sugiura M. The 2005 version of the chloroplast DNA sequence from tobacco (Nicotiana tabacum) Plant Mol Biol Rep. 2005;23(4):359–365.
-
- Tanaka M, Obokata J, Chunwongse J, Shinozaki K, Sugiura M. Rapid splicing and stepwise processing of a transcript from the psbB operon in tobacco chloroplasts: Determination of the intron sites in petB and petD. Mol Gen Genet. 1987;209(3):427–431. - PubMed
-
- Westhoff P, Herrmann RG. Complex RNA maturation in chloroplasts. The psbB operon from spinach. Eur J Biochem. 1988;171(3):551–564. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

