UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota
- PMID: 23509275
- PMCID: PMC3619370
- DOI: 10.1073/pnas.1303090110
UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota
Abstract
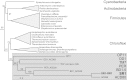
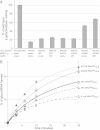
The composition of the human microbiota is recognized as an important factor in human health and disease. Many of our cohabitating microbes belong to phylum-level divisions for which there are no cultivated representatives and are only represented by small subunit rRNA sequences. For one such taxon (SR1), which includes bacteria with elevated abundance in periodontitis, we provide a single-cell genome sequence from a healthy oral sample. SR1 bacteria use a unique genetic code. In-frame TGA (opal) codons are found in most genes (85%), often at loci normally encoding conserved glycine residues. UGA appears not to function as a stop codon and is in equilibrium with the canonical GGN glycine codons, displaying strain-specific variation across the human population. SR1 encodes a divergent tRNA(Gly)UCA with an opal-decoding anticodon. SR1 glycyl-tRNA synthetase acylates tRNA(Gly)UCA with glycine in vitro with similar activity compared with normal tRNA(Gly)UCC. Coexpression of SR1 glycyl-tRNA synthetase and tRNA(Gly)UCA in Escherichia coli yields significant β-galactosidase activity in vivo from a lacZ gene containing an in-frame TGA codon. Comparative genomic analysis with Human Microbiome Project data revealed that the human body harbors a striking diversity of SR1 bacteria. This is a surprising finding because SR1 is most closely related to bacteria that live in anoxic and thermal environments. Some of these bacteria share common genetic and metabolic features with SR1, including UGA to glycine reassignment and an archaeal-type ribulose-1,5-bisphosphate carboxylase (RubisCO) involved in AMP recycling. UGA codon reassignment renders SR1 genes untranslatable by other bacteria, which impacts horizontal gene transfer within the human microbiota.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Perner M, et al. Microbial CO(2) fixation and sulfur cycling associated with low-temperature emissions at the Lilliput hydrothermal field, southern Mid-Atlantic Ridge (9° S) Environ Microbiol. 2007;9(5):1186–1201. - PubMed
-
- Hongoh Y, Ohkuma M, Kudo T. Molecular analysis of bacterial microbiota in the gut of the termite Reticulitermes speratus (Isoptera; Rhinotermitidae) FEMS Microbiol Ecol. 2003;44(2):231–242. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

