Met synergizes with p53 loss to induce mammary tumors that possess features of claudin-low breast cancer
- PMID: 23509284
- PMCID: PMC3619286
- DOI: 10.1073/pnas.1210353110
Met synergizes with p53 loss to induce mammary tumors that possess features of claudin-low breast cancer
Abstract
Triple-negative breast cancer (TNBC) accounts for ∼20% of cases and contributes to basal and claudin-low molecular subclasses of the disease. TNBCs have poor prognosis, display frequent mutations in tumor suppressor gene p53 (TP53), and lack targeted therapies. The MET receptor tyrosine kinase is elevated in TNBC and transgenic Met models (Met(mt)) develop basal-like tumors. To investigate collaborating events in the genesis of TNBC, we generated Met(mt) mice with conditional loss of murine p53 (Trp53) in mammary epithelia. Somatic Trp53 loss, in combination with Met(mt), significantly increased tumor penetrance over Met(mt) or Trp53 loss alone. Unlike Met(mt) tumors, which are histologically diverse and enriched in a basal-like molecular signature, the majority of Met(mt) tumors with Trp53 loss displayed a spindloid pathology with a distinct molecular signature that resembles the human claudin-low subtype of TNBC, including diminished claudins, an epithelial-to-mesenchymal transition signature, and decreased expression of the microRNA-200 family. Moreover, although mammary specific loss of Trp53 promotes tumors with diverse pathologies, those with spindloid pathology and claudin-low signature display genomic Met amplification. In both models, MET activity is required for maintenance of the claudin-low morphological phenotype, in which MET inhibitors restore cell-cell junctions, rescue claudin 1 expression, and abrogate growth and dissemination of cells in vivo. Among human breast cancers, elevated levels of MET and stabilized TP53, indicative of mutation, correlate with highly proliferative TNBCs of poor outcome. This work shows synergy between MET and TP53 loss for claudin-low breast cancer, identifies a restricted claudin-low gene signature, and provides a rationale for anti-MET therapies in TNBC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
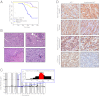
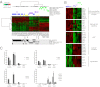
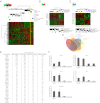
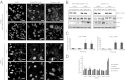



References
-
- Ferlay, et al. Globocan 2008 v2.0, Cancer Incidence and Mortality Worldwide. IARC CancerBase 10. 2008. Available at https://globocan.iarc.fr.
-
- Brenton JD, Carey LA, Ahmed AA, Caldas C. Molecular classification and molecular forecasting of breast cancer: Ready for clinical application? J Clin Oncol. 2005;23(29):7350–7360. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

