Identification of a previously undescribed divergent virus from the Flaviviridae family in an outbreak of equine serum hepatitis
- PMID: 23509292
- PMCID: PMC3625295
- DOI: 10.1073/pnas.1219217110
Identification of a previously undescribed divergent virus from the Flaviviridae family in an outbreak of equine serum hepatitis
Abstract
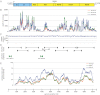
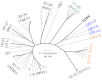
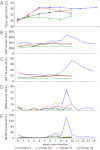
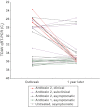
Theiler's disease is an acute hepatitis in horses that is associated with the administration of equine blood products; its etiologic agent has remained unknown for nearly a century. Here, we used massively parallel sequencing to explore samples from a recent Theiler's disease outbreak. Metatranscriptomic analysis of the short sequence reads identified a 10.5-kb sequence from a previously undescribed virus of the Flaviviridae family, which we designate "Theiler's disease-associated virus" (TDAV). Phylogenetic analysis clusters TDAV with GB viruses of the recently proposed Pegivirus genus, although it shares only 35.3% amino acid identity with its closest relative, GB virus D. An epidemiological survey of additional horses from three separate locations supports an association between TDAV infection and acute serum hepatitis. Experimental inoculation of horses with TDAV-positive plasma provides evidence that several weeks of viremia preceded liver injury and that liver disease may not be directly related to the level of viremia. Like hepatitis C virus, the best characterized Flaviviridae species known to cause hepatitis, we find TDAV is capable of efficient parenteral transmission, engendering acute and chronic infections associated with a diversity of clinical presentations ranging from subclinical infection to clinical hepatitis.
Conflict of interest statement
Conflict of interest statement: A.L.K., S.C., P.S.-C., T.J.D., and B.C.T. are coinventors on a patent application relating to the results herein. A.L.K., S.C., P.S.-C., D.E.G., and W.Z. are employees of the Novartis Institutes for BioMedical Research.
Figures



References
-
- Kahn CM, Line S, editors. 2011. Merck Veterinary Manual (Merck, Whitehouse Station, NJ), 9th Ed. Available at www.merckvetmanual.com/mvm/index.jsp?cfile=htm/bc/22802.htm.
-
- Theiler A. 1919. Acute liver-atrophy and parenchymatous hepatitis in horses. The Fifth and Sixth Reports of the Director of Veterinary Research, April, 1918. Department of Agriculture, Union of South Africa (The Government Printing and Stationery Office, Pretoria, Union of South Africa), pp. 7–164.
-
- Panciera RJ. Serum hepatitis in the horse. J Am Vet Med Assoc. 1969;155(2):408–410. - PubMed
-
- Hjerpe CA. Serum hepatitis in the horse. J Am Vet Med Assoc. 1964;144:734–740. - PubMed
-
- Aleman M, Nieto JE, Carr EA, Carlson GP. Serum hepatitis associated with commercial plasma transfusion in horses. J Vet Intern Med. 2005;19(1):120–122. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

