RNase P-associated external guide sequence effectively reduces the expression of human CC-chemokine receptor 5 and inhibits the infection of human immunodeficiency virus 1
- PMID: 23509733
- PMCID: PMC3591226
- DOI: 10.1155/2013/509714
RNase P-associated external guide sequence effectively reduces the expression of human CC-chemokine receptor 5 and inhibits the infection of human immunodeficiency virus 1
Abstract
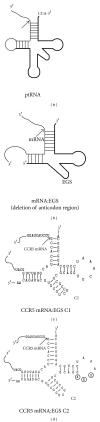
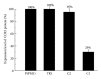
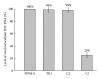
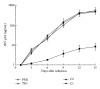
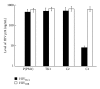
External guide sequences (EGSs) represent a new class of RNA-based gene-targeting agents, consist of a sequence complementary to a target mRNA, and render the target RNA susceptible to degradation by ribonuclease P (RNase P). In this study, EGSs were constructed to target the mRNA encoding human CC-chemokine receptor 5 (CCR5), one of the primary coreceptors for HIV. An EGS RNA, C1, efficiently directed human RNase P to cleave the CCR5 mRNA sequence in vitro. A reduction of about 70% in the expression level of both CCR5 mRNA and protein and an inhibition of more than 50-fold in HIV (R5 strain Ba-L) p24 production were observed in cells that expressed C1. In comparison, a reduction of about 10% in the expression of CCR5 and viral growth was found in cells that either did not express the EGS or produced a "disabled" EGS which carried nucleotide mutations that precluded RNase P recognition. Furthermore, the same C1-expressing cells that were protected from R5 strain Ba-L retained susceptibility to X4 strain IIIB, which uses CXCR4 as the coreceptor instead of CCR5, suggesting that the RNase P-mediated cleavage induced by the EGS is specific for the target CCR5 but not the closely related CXCR4. Our results provide direct evidence that EGS RNAs against CCR5 are effective and specific in blocking HIV infection and growth. These results also demonstrate the feasibility to develop highly effective EGSs for anti-HIV therapy.
Figures



References
-
- Scherer LJ, Rossi JJ. Approaches for the sequence-specific knockdown of mRNA. Nature Biotechnology. 2003;21(12):1457–1465. - PubMed
-
- Stein CA, Cheng YC. Antisense oligonucleotides as therapeutic agents—is the bullet really magical? Science. 1993;261(5124):1004–1012. - PubMed
-
- Wong-Staal F, Poeschla EM, Looney DJ. A controlled, Phase 1 clinical trial to evaluate the safety and effects in HIV-1 infected humans of autologous lymphocytes transduced with a ribozyme that cleaves HIV-1 RNA. Human Gene Therapy. 1998;9(16):2407–2425. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

