Measurement and modeling of intrinsic transcription terminators
- PMID: 23511967
- PMCID: PMC3643576
- DOI: 10.1093/nar/gkt163
Measurement and modeling of intrinsic transcription terminators
Erratum in
-
Measurement and modeling of intrinsic transcription terminators.Nucleic Acids Res. 2016 Aug 19;44(14):7006. doi: 10.1093/nar/gkw379. Epub 2016 Apr 29. Nucleic Acids Res. 2016. PMID: 27131373 Free PMC article. No abstract available.
Abstract
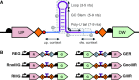
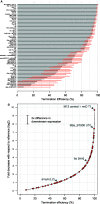
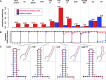
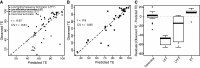
The reliable forward engineering of genetic systems remains limited by the ad hoc reuse of many types of basic genetic elements. Although a few intrinsic prokaryotic transcription terminators are used routinely, termination efficiencies have not been studied systematically. Here, we developed and validated a genetic architecture that enables reliable measurement of termination efficiencies. We then assembled a collection of 61 natural and synthetic terminators that collectively encode termination efficiencies across an ∼800-fold dynamic range within Escherichia coli. We simulated co-transcriptional RNA folding dynamics to identify competing secondary structures that might interfere with terminator folding kinetics or impact termination activity. We found that structures extending beyond the core terminator stem are likely to increase terminator activity. By excluding terminators encoding such context-confounding elements, we were able to develop a linear sequence-function model that can be used to estimate termination efficiencies (r = 0.9, n = 31) better than models trained on all terminators (r = 0.67, n = 54). The resulting systematically measured collection of terminators should improve the engineering of synthetic genetic systems and also advance quantitative modeling of transcription termination.
Figures