What do we know about IDH1/2 mutations so far, and how do we use it?
- PMID: 23512379
- PMCID: PMC3633675
- DOI: 10.1007/s00401-013-1106-9
What do we know about IDH1/2 mutations so far, and how do we use it?
Abstract
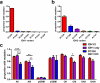
Whole genome analyses have facilitated the discovery of clinically relevant genetic alterations in a variety of diseases, most notably cancer. A prominent example of this was the discovery of mutations in isocitrate dehydrogenases 1 and 2 (IDH1/2) in a sizeable proportion of gliomas and some other neoplasms. Herein the normal functions of these enzymes, how the mutations alter their catalytic properties, the effects of their D-2-hydroxyglutarate metabolite, technical considerations in diagnostic neuropathology, implications about prognosis and therapeutic considerations, and practical applications and controversies regarding IDH1/2 mutation testing are discussed.
Figures


References
-
- Abbas S, Lugthart S, Kavelaars FG, et al. Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: prevalence and prognostic value. Blood. 2010;116(12):2122–6. - PubMed
-
- Aghili M, Zahedi F, Rafiee E. Hydroxyglutaric aciduria and malignant brain tumor: a case report and literature review. J Neurooncol. 2009;91:233–6. - PubMed
-
- Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9(6):1191–200. - PubMed
-
- Ahmadi R, Stockhammer F, Becker N, et al. No prognostic value of IDH1 mutations in a series of 100 WHO grade II astrocytomas. J Neurooncol. 2012;109(1):15–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

