Single-cell imaging of HIV-1 provirus (SCIP)
- PMID: 23513220
- PMCID: PMC3619299
- DOI: 10.1073/pnas.1216254110
Single-cell imaging of HIV-1 provirus (SCIP)
Abstract
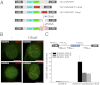
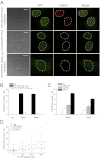
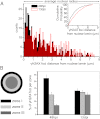
Recent advances in fluorescence microscopy provided tools for the investigation and the analysis of the viral replication steps in the cellular context. In the HIV field, the current visualization systems successfully achieve the fluorescent labeling of the viral envelope and proteins, but not the genome. Here, we developed a system able to visualize the proviral DNA of HIV-1 through immunofluorescence detection of repair foci for DNA double-strand breaks specifically induced in the viral genome by the heterologous expression of the I-SceI endonuclease. The system for Single-Cell Imaging of HIV-1 Provirus, named SCIP, provides the possibility to individually track integrated-viral DNA within the nuclei of infected cells. In particular, SCIP allowed us to perform a topological analysis of integrated viral DNA revealing that HIV-1 preferentially integrates in the chromatin localized at the periphery of the nuclei.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

