Lipid-induced conformational changes within the cytochrome b6f complex of oxygenic photosynthesis
- PMID: 23514009
- PMCID: PMC4034689
- DOI: 10.1021/bi301638h
Lipid-induced conformational changes within the cytochrome b6f complex of oxygenic photosynthesis
Abstract
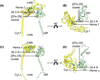
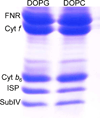
Cytochrome b6f catalyzes quinone redox reactions within photosynthetic membranes to generate a transmembrane proton electrochemical gradient for ATP synthesis. A key step involves the transfer of an electron from the [2Fe-2S] cluster of the iron-sulfur protein (ISP) extrinsic domain to the cytochrome f heme across a distance of 26 Å, which is too large for competent electron transfer but could be bridged by translation-rotation of the ISP. Here we report the first crystallographic evidence of significant motion of the ISP extrinsic domain. It is inferred that extensive crystallographic disorder of the ISP extrinsic domain indicates conformational flexibility. The ISP disorder observed in this structure, in contrast to the largely ordered ISP structure observed in the b6f complex supplemented with neutral lipids, is attributed to electrostatic interactions arising from anionic lipids.
Figures
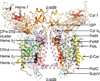

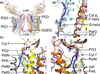
References
-
- Schutz M, Brugna M, Lebrun E, Baymann F, Huber R, Stetter KO, Hauska G, Toci R, Lemesle-Meunier D, Tron P, Schmidt C, Nitschke W. Early evolution of cytochrome bc complexes. Journal of molecular biology. 2000;300:663–675. - PubMed
-
- Darrouzet E, Cooley JW, Daldal F. The Cytochrome bc (1) Complex and its Homologue the b (6) f Complex: Similarities and Differences. Photosynth Res. 2004;79:25–44. - PubMed
-
- Widger WR, Cramer WA, Herrmann RG, Trebst A. Sequence homology and structural similarity between cytochrome b of mitochondrial complex III and the chloroplast b6-f complex: position of the cytochrome b hemes in the membrane. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:674–678. - PMC - PubMed
-
- Berry EA, Guergova-Kuras M, Huang LS, Crofts AR. Structure and function of cytochrome bc complexes. Annual review of biochemistry. 2000;69:1005–1075. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

