The Enzyme Portal: a case study in applying user-centred design methods in bioinformatics
- PMID: 23514033
- PMCID: PMC3623738
- DOI: 10.1186/1471-2105-14-103
The Enzyme Portal: a case study in applying user-centred design methods in bioinformatics
Abstract
User-centred design (UCD) is a type of user interface design in which the needs and desires of users are taken into account at each stage of the design process for a service or product; often for software applications and websites. Its goal is to facilitate the design of software that is both useful and easy to use. To achieve this, you must characterise users' requirements, design suitable interactions to meet their needs, and test your designs using prototypes and real life scenarios.For bioinformatics, there is little practical information available regarding how to carry out UCD in practice. To address this we describe a complete, multi-stage UCD process used for creating a new bioinformatics resource for integrating enzyme information, called the Enzyme Portal (http://www.ebi.ac.uk/enzymeportal). This freely-available service mines and displays data about proteins with enzymatic activity from public repositories via a single search, and includes biochemical reactions, biological pathways, small molecule chemistry, disease information, 3D protein structures and relevant scientific literature.We employed several UCD techniques, including: persona development, interviews, 'canvas sort' card sorting, user workflows, usability testing and others. Our hope is that this case study will motivate the reader to apply similar UCD approaches to their own software design for bioinformatics. Indeed, we found the benefits included more effective decision-making for design ideas and technologies; enhanced team-working and communication; cost effectiveness; and ultimately a service that more closely meets the needs of our target audience.
Figures
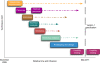







References
-
- Aurrecoechea C, Brestelli J, Brunk BP, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, Innamorato F, Iodice J, Kissinger JC, Kraemer ET, Li W, Nayak V, Pennington C, Pinney DF, Roos DS, Ross C, Srinivasamoorthy G, Stoeckert CJ, Thibodeau R, Treatman C, Wang H. Miller J a. EuPathDB: a portal to eukaryotic pathogen databases. Nucleic Acids Res. 2010;38:D415–9. doi: 10.1093/nar/gkp941. - DOI - PMC - PubMed
-
- Ringwald M, Iyer V, Mason JC, Stone KR, Tadepally HD, Bult CJ, Eppig JT, Oakley DJ, Briois S, Stupka E, Maselli V, Smedley D, Liu S, Hansen J, Baldock R, Hicks GG, Skarnes WC. Kadin J a. The IKMC web portal: a central point of entry to data and resources from the International Knockout Mouse Consortium. Nucleic Acids Res. 2011;39:D849–55. doi: 10.1093/nar/gkq879. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

