Evaluation and integration of existing methods for computational prediction of allergens
- PMID: 23514097
- PMCID: PMC3599076
- DOI: 10.1186/1471-2105-14-S4-S1
Evaluation and integration of existing methods for computational prediction of allergens
Abstract
Background: Allergy involves a series of complex reactions and factors that contribute to the development of the disease and triggering of the symptoms, including rhinitis, asthma, atopic eczema, skin sensitivity, even acute and fatal anaphylactic shock. Prediction and evaluation of the potential allergenicity is of importance for safety evaluation of foods and other environment factors. Although several computational approaches for assessing the potential allergenicity of proteins have been developed, their performance and relative merits and shortcomings have not been compared systematically.
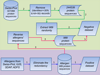
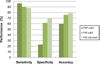
Results: To evaluate and improve the existing methods for allergen prediction, we collected an up-to-date definitive dataset consisting of 989 known allergens and massive putative non-allergens. The three most widely used allergen computational prediction approaches including sequence-, motif- and SVM-based (Support Vector Machine) methods were systematically compared using the defined parameters and we found that SVM-based method outperformed the other two methods with higher accuracy and specificity. The sequence-based method with the criteria defined by FAO/WHO (FAO: Food and Agriculture Organization of the United Nations; WHO: World Health Organization) has higher sensitivity of over 98%, but having a low specificity. The advantage of motif-based method is the ability to visualize the key motif within the allergen. Notably, the performances of the sequence-based method defined by FAO/WHO and motif eliciting strategy could be improved by the optimization of parameters. To facilitate the allergen prediction, we integrated these three methods in a web-based application proAP, which provides the global search of the known allergens and a powerful tool for allergen predication. Flexible parameter setting and batch prediction were also implemented. The proAP can be accessed at http://gmobl.sjtu.edu.cn/proAP/main.html.
Conclusions: This study comprehensively evaluated sequence-, motif- and SVM-based computational prediction approaches for allergens and optimized their parameters to obtain better performance. These findings may provide helpful guidance for the researchers in allergen-prediction. Furthermore, we integrated these methods into a web application proAP, greatly facilitating users to do customizable allergen search and prediction.
Figures




Similar articles
-
AllerTOP--a server for in silico prediction of allergens.BMC Bioinformatics. 2013;14 Suppl 6(Suppl 6):S4. doi: 10.1186/1471-2105-14-S6-S4. Epub 2013 Apr 17. BMC Bioinformatics. 2013. PMID: 23735058 Free PMC article.
-
Improving Allergen Prediction in Main Crops Using a Weighted Integrative Method.Interdiscip Sci. 2017 Dec;9(4):545-549. doi: 10.1007/s12539-016-0192-5. Epub 2016 Oct 12. Interdiscip Sci. 2017. PMID: 27734271
-
[Major revision of the allergen database for food safety (ADFS) and validation of the motif-based allergenicity prediction tool].Kokuritsu Iyakuhin Shokuhin Eisei Kenkyusho Hokoku. 2009;(127):44-9. Kokuritsu Iyakuhin Shokuhin Eisei Kenkyusho Hokoku. 2009. PMID: 20306706 Japanese.
-
Practical and predictive bioinformatics methods for the identification of potentially cross-reactive protein matches.Mol Nutr Food Res. 2006 Jul;50(7):655-60. doi: 10.1002/mnfr.200500277. Mol Nutr Food Res. 2006. PMID: 16810734 Review.
-
Bioinformatics approaches to classifying allergens and predicting cross-reactivity.Immunol Allergy Clin North Am. 2007 Feb;27(1):1-27. doi: 10.1016/j.iac.2006.11.005. Immunol Allergy Clin North Am. 2007. PMID: 17276876 Free PMC article. Review.
Cited by
-
Pathway and network approaches for identification of cancer signature markers from omics data.J Cancer. 2015 Jan 1;6(1):54-65. doi: 10.7150/jca.10631. eCollection 2015. J Cancer. 2015. PMID: 25553089 Free PMC article. Review.
-
Peptide Property Prediction for Mass Spectrometry Using AI: An Introduction to State of the Art Models.Proteomics. 2025 May;25(9-10):e202400398. doi: 10.1002/pmic.202400398. Epub 2025 Apr 10. Proteomics. 2025. PMID: 40211610 Free PMC article. Review.
-
PREAL: prediction of allergenic protein by maximum Relevance Minimum Redundancy (mRMR) feature selection.BMC Syst Biol. 2013;7 Suppl 5(Suppl 5):S9. doi: 10.1186/1752-0509-7-S5-S9. Epub 2013 Dec 9. BMC Syst Biol. 2013. PMID: 24565053 Free PMC article.
-
Current and prospective computational approaches and challenges for developing COVID-19 vaccines.Adv Drug Deliv Rev. 2021 May;172:249-274. doi: 10.1016/j.addr.2021.02.004. Epub 2021 Feb 6. Adv Drug Deliv Rev. 2021. PMID: 33561453 Free PMC article. Review.
-
Computational approaches for molecular characterization and structure-based functional elucidation of a hypothetical protein from Mycobacterium tuberculosis.Genomics Inform. 2023 Jun;21(2):e25. doi: 10.5808/gi.23001. Epub 2023 Jun 30. Genomics Inform. 2023. PMID: 37415455 Free PMC article.
References
-
- Mekori YA. Introduction to allergic diseases. Crit Rev Food Sci Nutr. 1996;36(Suppl.):S1–S18. - PubMed
-
- Metcalfe DD, Astwood JD, Townsend R, Sampson HA, Taylor SL, Fuchs RL. Assessment of the allergenic potential of foods derived from genetically engineered crop plants. Crit Rev Food Sci Nutr. 1996;36(Suppl.):S165–S186. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

