Structural insights into the regulation of foreign genes in Salmonella by the Hha/H-NS complex
- PMID: 23515315
- PMCID: PMC3650374
- DOI: 10.1074/jbc.M113.455378
Structural insights into the regulation of foreign genes in Salmonella by the Hha/H-NS complex
Abstract
Background: Hha facilitates H-NS-mediated silencing of foreign genes in bacteria.
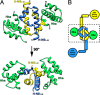
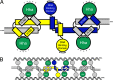
Results: Two Hha monomers bind opposing faces of the H-NS N-terminal dimerization domain.
Conclusion: Hha binds the dimerization domain of H-NS and may contact DNA via positively charged surface residues.
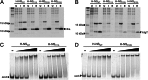
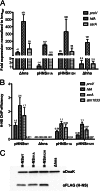
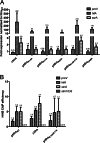
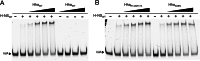
Significance: The structure of Hha and H-NS in complex provides a mechanistic model of how Hha may affect gene regulation. The bacterial nucleoid-associated proteins Hha and H-NS jointly repress horizontally acquired genes in Salmonella, including essential virulence loci encoded within Salmonella pathogenicity islands. Hha is known to interact with the N-terminal dimerization domain of H-NS; however, the manner in which this interaction enhances transcriptional silencing is not understood. To further understand this process, we solved the x-ray crystal structure of Hha in complex with the N-terminal dimerization domain of H-NS (H-NS(1-46)) to 3.2 Å resolution. Two monomers of Hha bind to symmetrical sites on either side of the H-NS(1-46) dimer. Disruption of the Hha/H-NS interaction by the H-NS site-specific mutation I11A results in increased expression of the Hha/H-NS co-regulated gene hilA without affecting the expression levels of proV, a target gene repressed by H-NS in an Hha-independent fashion. Examination of the structure revealed a cluster of conserved basic amino acids that protrude from the surface of Hha on the opposite side of the Hha/H-NS(1-46) interface. Hha mutants with a diminished positively charged surface maintain the ability to interact with H-NS but can no longer regulate hilA. Increased expression of the hilA locus did not correspond to significant depletion of H-NS at the promoter region in chromatin immunoprecipitation assays. However, in vitro, we find Hha improves H-NS binding to target DNA fragments. Taken together, our results show for the first time how Hha and H-NS interact to direct transcriptional repression and reveal that a positively charged surface of Hha enhances the silencing activity of H-NS nucleoprotein filaments.
Keywords: Bacterial Genetics; DNA-binding Protein; Gene Regulation; Gene Transfer; Microbial Pathogenesis; Microbiology.
Figures


References
-
- Lawrence J. G., Ochman H. (1997) Amelioration of bacterial genomes. Rates of change and exchange. J. Mol. Evol. 44, 383–397 - PubMed
-
- Groisman E. A., Ochman H. (1996) Pathogenicity islands. Bacterial evolution in quantum leaps. Cell 87, 791–794 - PubMed
-
- Navarre W. W., Porwollik S., Wang Y., McClelland M., Rosen H., Libby S. J., Fang F. C. (2006) Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313, 236–238 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

