Molecular dialogue between the human gut microbiota and the host: a Lactobacillus and Bifidobacterium perspective
- PMID: 23516017
- PMCID: PMC11113728
- DOI: 10.1007/s00018-013-1318-0
Molecular dialogue between the human gut microbiota and the host: a Lactobacillus and Bifidobacterium perspective
Abstract
The human gut represents a highly complex ecosystem, which is densely colonized by a myriad of microorganisms that influence the physiology, immune function and health status of the host. Among the many members of the human gut microbiota, there are microorganisms that have co-evolved with their host and that are believed to exert health-promoting or probiotic effects. Probiotic bacteria isolated from the gut and other environments are commercially exploited, and although there is a growing list of health benefits provided by the consumption of such probiotics, their precise mechanisms of action have essentially remained elusive. Genomics approaches have provided exciting new opportunities for the identification of probiotic effector molecules that elicit specific responses to influence the physiology and immune function of their human host. In this review, we describe the current understanding of the intriguing relationships that exist between the human gut and key members of the gut microbiota such as bifidobacteria and lactobacilli, discussed here as prototypical groups of probiotic microorganisms.
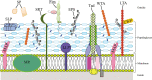
Figures

References
-
- Marco ML, Pavan S, Kleerebezem M. Towards understanding molecular modes of probiotic action. Curr Opin Biotechnol. 2006;17(2):204–210. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

