Chaste: an open source C++ library for computational physiology and biology
- PMID: 23516352
- PMCID: PMC3597547
- DOI: 10.1371/journal.pcbi.1002970
Chaste: an open source C++ library for computational physiology and biology
Abstract
Chaste - Cancer, Heart And Soft Tissue Environment - is an open source C++ library for the computational simulation of mathematical models developed for physiology and biology. Code development has been driven by two initial applications: cardiac electrophysiology and cancer development. A large number of cardiac electrophysiology studies have been enabled and performed, including high-performance computational investigations of defibrillation on realistic human cardiac geometries. New models for the initiation and growth of tumours have been developed. In particular, cell-based simulations have provided novel insight into the role of stem cells in the colorectal crypt. Chaste is constantly evolving and is now being applied to a far wider range of problems. The code provides modules for handling common scientific computing components, such as meshes and solvers for ordinary and partial differential equations (ODEs/PDEs). Re-use of these components avoids the need for researchers to 're-invent the wheel' with each new project, accelerating the rate of progress in new applications. Chaste is developed using industrially-derived techniques, in particular test-driven development, to ensure code quality, re-use and reliability. In this article we provide examples that illustrate the types of problems Chaste can be used to solve, which can be run on a desktop computer. We highlight some scientific studies that have used or are using Chaste, and the insights they have provided. The source code, both for specific releases and the development version, is available to download under an open source Berkeley Software Distribution (BSD) licence at http://www.cs.ox.ac.uk/chaste, together with details of a mailing list and links to documentation and tutorials.
Conflict of interest statement
I have read the journal's policy and have the following conflicts: GRM and DJG have received research support from GlaxoSmithKline Plc.
Figures
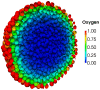

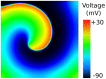
 monodomain simulation using the Luo-Rudy 1991 action-potential model with the modifications and protocol suggested in . See also Video S3.
monodomain simulation using the Luo-Rudy 1991 action-potential model with the modifications and protocol suggested in . See also Video S3.
References
-
- Wilson G (2006) Where's the real bottleneck in scientific computing? American Scientist 94: 5–6.
-
- Cooper J, Cervenansky F, De Fabritiis G, Fenner J, Friboulet D, et al. (2010) The virtual physiological human toolkit. Phil Trans R Soc A 368: 3925–3936. - PubMed
-
- Merali Z (2010) Computational science: Error, why scientific programming does not compute. Nature 467: 775–777. - PubMed
-
- Le Nov'ere N, Finney A, Hucka M, Bhalla US, Campagne F, et al. (2005) Minimum information requested in the annotation of biochemical models (MIRIAM). Nat Biotech 23: 1509–1515. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

