Trichoderma-plant root colonization: escaping early plant defense responses and activation of the antioxidant machinery for saline stress tolerance
- PMID: 23516362
- PMCID: PMC3597500
- DOI: 10.1371/journal.ppat.1003221
Trichoderma-plant root colonization: escaping early plant defense responses and activation of the antioxidant machinery for saline stress tolerance
Erratum in
- PLoS Pathog. 2013 Apr;9(4). doi:10.1371/annotation/8b818c15-3fe0-4e56-9be2-e44fd1ed3fae. Takayuki, Tohge [corrected to Tohge, Takayuki]
Abstract
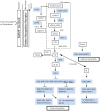
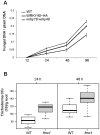
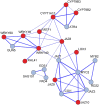
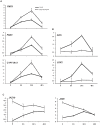
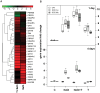
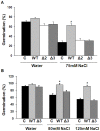
Trichoderma spp. are versatile opportunistic plant symbionts which can colonize the apoplast of plant roots. Microarrays analysis of Arabidopsis thaliana roots inoculated with Trichoderma asperelloides T203, coupled with qPCR analysis of 137 stress responsive genes and transcription factors, revealed wide gene transcript reprogramming, proceeded by a transient repression of the plant immune responses supposedly to allow root colonization. Enhancement in the expression of WRKY18 and WRKY40, which stimulate JA-signaling via suppression of JAZ repressors and negatively regulate the expression of the defense genes FMO1, PAD3 and CYP71A13, was detected in Arabidopsis roots upon Trichoderma colonization. Reduced root colonization was observed in the wrky18/wrky40 double mutant line, while partial phenotypic complementation was achieved by over-expressing WRKY40 in the wrky18 wrky40 background. On the other hand increased colonization rate was found in roots of the FMO1 knockout mutant. Trichoderma spp. stimulate plant growth and resistance to a wide range of adverse environmental conditions. Arabidopsis and cucumber (Cucumis sativus L.) plants treated with Trichoderma prior to salt stress imposition show significantly improved seed germination. In addition, Trichoderma treatment affects the expression of several genes related to osmo-protection and general oxidative stress in roots of both plants. The MDAR gene coding for monodehydroascorbate reductase is significantly up-regulated and, accordingly, the pool of reduced ascorbic acid was found to be increased in Trichoderma treated plants. 1-Aminocyclopropane-1-carboxylate (ACC)-deaminase silenced Trichoderma mutants were less effective in providing tolerance to salt stress, suggesting that Trichoderma, similarly to ACC deaminase producing bacteria, can ameliorate plant growth under conditions of abiotic stress, by lowering ameliorating increases in ethylene levels as well as promoting an elevated antioxidative capacity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Brotman Y, Kapuganti JG, Viterbo A (2010) Trichoderma . Curr Biol 20: R390–391. - PubMed
-
- Lorito M, Woo SL, Harman GE, Monte E (2010) Translational Research on Trichoderma: From 'Omics to the Field. Annu Rev Phytopathol 48: 395–417. - PubMed
-
- Viterbo A, & Horwitz BA (2010) Mycoparasitism. In: Ebbole KABDJ, editor. Cellular and Molecular Biology of Filamentous Fungi. Washington: American Society for Microbiology. pp. 676–693.
-
- Shoresh M, Harman GE, Mastouri F (2010) Induced systemic resistance and plant responses to fungal biocontrol agents. Annu Rev Phytopathol 48: 21–43. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

