Rare copy number variants are a common cause of short stature
- PMID: 23516380
- PMCID: PMC3597495
- DOI: 10.1371/journal.pgen.1003365
Rare copy number variants are a common cause of short stature
Abstract
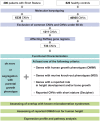
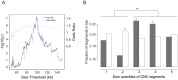
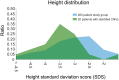
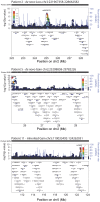
Human growth has an estimated heritability of about 80%-90%. Nevertheless, the underlying cause of shortness of stature remains unknown in the majority of individuals. Genome-wide association studies (GWAS) showed that both common single nucleotide polymorphisms and copy number variants (CNVs) contribute to height variation under a polygenic model, although explaining only a small fraction of overall genetic variability in the general population. Under the hypothesis that severe forms of growth retardation might also be caused by major gene effects, we searched for rare CNVs in 200 families, 92 sporadic and 108 familial, with idiopathic short stature compared to 820 control individuals. Although similar in number, patients had overall significantly larger CNVs (p-value<1×10(-7)). In a gene-based analysis of all non-polymorphic CNVs>50 kb for gene function, tissue expression, and murine knock-out phenotypes, we identified 10 duplications and 10 deletions ranging in size from 109 kb to 14 Mb, of which 7 were de novo (p<0.03) and 13 inherited from the likewise affected parent but absent in controls. Patients with these likely disease causing 20 CNVs were smaller than the remaining group (p<0.01). Eleven (55%) of these CNVs either overlapped with known microaberration syndromes associated with short stature or contained GWAS loci for height. Haploinsufficiency (HI) score and further expression profiling suggested dosage sensitivity of major growth-related genes at these loci. Overall 10% of patients carried a disease-causing CNV indicating that, like in neurodevelopmental disorders, rare CNVs are a frequent cause of severe growth retardation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Visscher PM, Hill WG, Wray NR (2008) Heritability in the genomics era–concepts and misconceptions. Nat Rev Genet 9: 255266. - PubMed
-
- Mendez H (1985) Introduction to the study of pre- and postnatal growth in humans: a review. Am J Med Genet 20: 63–85. - PubMed
-
- Superti-Furga A, Bonafe L, Rimoin DL (2001) Molecular-pathogenetic classification of genetic disorders of the skeleton. Am J Med Genet 106: 282–293. - PubMed
-
- Kallen B, Knudsen LB, Mutchinick O, Mastroiacovo P, Lancaster P, et al. (1993) Monitoring dominant germ cell mutations using skeletal dysplasias registered in malformation registries: an international feasibility study. Int J Epidemiol 22: 107–115. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

