A conserved upstream motif orchestrates autonomous, germline-enriched expression of Caenorhabditis elegans piRNAs
- PMID: 23516384
- PMCID: PMC3597512
- DOI: 10.1371/journal.pgen.1003392
A conserved upstream motif orchestrates autonomous, germline-enriched expression of Caenorhabditis elegans piRNAs
Abstract
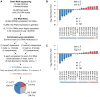
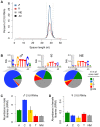
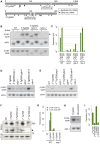
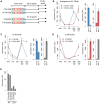
Piwi-interacting RNAs (piRNAs) fulfill a critical, conserved role in defending the genome against foreign genetic elements. In many organisms, piRNAs appear to be derived from processing of a long, polycistronic RNA precursor. Here, we establish that each Caenorhabditis elegans piRNA represents a tiny, autonomous transcriptional unit. Remarkably, the minimal C. elegans piRNA cassette requires only a 21 nucleotide (nt) piRNA sequence and an ∼50 nt upstream motif with limited genomic context for expression. Combining computational analyses with a novel, in vivo transgenic system, we demonstrate that this upstream motif is necessary for independent expression of a germline-enriched, Piwi-dependent piRNA. We further show that a single nucleotide position within this motif directs differential germline enrichment. Accordingly, over 70% of C. elegans piRNAs are selectively expressed in male or female germline, and comparison of the genes they target suggests that these two populations have evolved independently. Together, our results indicate that C. elegans piRNA upstream motifs act as independent promoters to specify which sequences are expressed as piRNAs, how abundantly they are expressed, and in what germline. As the genome encodes well over 15,000 unique piRNA sequences, our study reveals that the number of transcriptional units encoding piRNAs rivals the number of mRNA coding genes in the C. elegans genome.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Comment in
-
Small RNAs: Defining piRNA expression.Nat Rev Genet. 2013 May;14(5):301. doi: 10.1038/nrg3474. Epub 2013 Apr 4. Nat Rev Genet. 2013. PMID: 23552218 No abstract available.
References
-
- Lin H, Spradling AC (1997) A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124: 2463–2476. - PubMed
-
- Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA (2002) Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell 110: 689–699. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

