Structural phylogenomics reveals gradual evolutionary replacement of abiotic chemistries by protein enzymes in purine metabolism
- PMID: 23516625
- PMCID: PMC3596326
- DOI: 10.1371/journal.pone.0059300
Structural phylogenomics reveals gradual evolutionary replacement of abiotic chemistries by protein enzymes in purine metabolism
Abstract
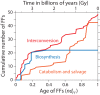
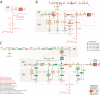
The origin of metabolism has been linked to abiotic chemistries that existed in our planet at the beginning of life. While plausible chemical pathways have been proposed, including the synthesis of nucleobases, ribose and ribonucleotides, the cooption of these reactions by modern enzymes remains shrouded in mystery. Here we study the emergence of purine metabolism. The ages of protein domains derived from a census of fold family structure in hundreds of genomes were mapped onto enzymes in metabolic diagrams. We find that the origin of the nucleotide interconversion pathway benefited most parsimoniously from the prebiotic formation of adenine nucleosides. In turn, pathways of nucleotide biosynthesis, catabolism and salvage originated ∼300 million years later by concerted enzymatic recruitments and gradual replacement of abiotic chemistries. Remarkably, this process led to the emergence of the fully enzymatic biosynthetic pathway ∼3 billion years ago, concurrently with the appearance of a functional ribosome. The simultaneous appearance of purine biosynthesis and the ribosome probably fulfilled the expanding matter-energy and processing needs of genomic information.
Conflict of interest statement
Figures


References
-
- Caetano-Anollés G, Yafremava LS, Gee H, Caetano-Anollés D, Mittenthal JE (2009) The origin and evolution of modern metabolism. Intl J Biochem Cell Biol 41: 285–297. - PubMed
-
- Caetano-Anollés G, Wang M, Caetano-Anollés D, Mittenthal JE (2009) The origin, evolution and structure of the protein world. Biochem J 417: 621–637. - PubMed
-
- Wang M, Caetano-Anollés G (2009) The evolutionary mechanics of domain organization in proteomes and the rise of modularity in the protein world. Structure 17: 66–78. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

