Antennal transcriptome analysis of the chemosensory gene families in the tree killing bark beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae)
- PMID: 23517120
- PMCID: PMC3610139
- DOI: 10.1186/1471-2164-14-198
Antennal transcriptome analysis of the chemosensory gene families in the tree killing bark beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae)
Abstract
Background: The European spruce bark beetle, Ips typographus, and the North American mountain pine beetle, Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae), are severe pests of coniferous forests. Both bark beetle species utilize aggregation pheromones to coordinate mass-attacks on host trees, while odorants from host and non-host trees modulate the pheromone response. Thus, the bark beetle olfactory sense is of utmost importance for fitness. However, information on the genes underlying olfactory detection has been lacking in bark beetles and is limited in Coleoptera. We assembled antennal transcriptomes from next-generation sequencing of I. typographus and D. ponderosae to identify members of the major chemosensory multi-gene families.
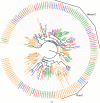
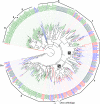
Results: Gene ontology (GO) annotation indicated that the relative abundance of transcripts associated with specific GO terms was highly similar in the two species. Transcripts with terms related to olfactory function were found in both species. Focusing on the chemosensory gene families, we identified 15 putative odorant binding proteins (OBP), 6 chemosensory proteins (CSP), 3 sensory neuron membrane proteins (SNMP), 43 odorant receptors (OR), 6 gustatory receptors (GR), and 7 ionotropic receptors (IR) in I. typographus; and 31 putative OBPs, 11 CSPs, 3 SNMPs, 49 ORs, 2 GRs, and 15 IRs in D. ponderosae. Predicted protein sequences were compared with counterparts in the flour beetle, Tribolium castaneum, the cerambycid beetle, Megacyllene caryae, and the fruit fly, Drosophila melanogaster. The most notable result was found among the ORs, for which large bark beetle-specific expansions were found. However, some clades contained receptors from all four beetle species, indicating a degree of conservation among some coleopteran OR lineages. Putative GRs for carbon dioxide and orthologues for the conserved antennal IRs were included in the identified receptor sets.
Conclusions: The protein families important for chemoreception have now been identified in three coleopteran species (four species for the ORs). Thus, this study allows for improved evolutionary analyses of coleopteran olfaction. Identification of these proteins in two of the most destructive forest pests, sharing many semiochemicals, is especially important as they might represent novel targets for population control.
Figures




References
-
- Schlyter F, Birgersson GA. In: Pheromones of non-Lepidopteran insects associated with agricultural plants. Hardie J, Minks AK, editor. CAB International, Oxford; 1999. Forest beetles; pp. 113–148.
-
- Zhang Q-H, Schlyter F. Olfactory recognition and behavioural avoidance of angiosperm nonhost volatiles by conifer-inhabiting bark beetles. Agric For Entomol. 2004;6(1):1–19. doi: 10.1111/j.1461-9555.2004.00202.x. - DOI
-
- Erbilgin N, Krokene P, Kvamme T, Christiansen E. A host monoterpene influences Ips typographus (Coleoptera: Curculionidae, Scolytinae) responses to its aggregation pheromone. Agric For Entomol. 2007;9:135–140. doi: 10.1111/j.1461-9563.2007.00329.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

