Development of porcine rotavirus vp6 protein based ELISA for differentiation of this virus and other viruses
- PMID: 23517810
- PMCID: PMC3658953
- DOI: 10.1186/1743-422X-10-91
Development of porcine rotavirus vp6 protein based ELISA for differentiation of this virus and other viruses
Abstract
Background: The context and purpose of the study included 1) bacterial expression of viral protein 6 (VP6) of porcine rotavirus (PRV) and generation of rabbit polyclonal antiserum to the VP6 protein; 3) establishment of a discrimination ELISA to distinguish PRV from a panel of other porcine viruses.
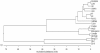
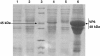
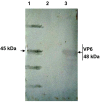
Results: The VP6 gene of PRV isolate DN30209 amplified by reverse transcription-PCR was 1356 bp containing a complete open reading frame (ORF) encoding 397 amino acids. Sequence comparison and phylogenetic analysis indicated that PRV DN30209 may belong to group A of rotavirus. Bacterially expressed VP6 was expressed in E.coli and anti-VP6 antibody was capable of distinguishing PRV from Porcine transmissible gastroenteritis virus, Porcine epidemic diarrhea virus, Porcine circovirus type II, Porcine reproductive and respiratory syndrome virus, Porcine pseudorabies virus and Porcine parvovirus.
Conclusions: PRV VP6 expressed in E. coli can be used to generate antibodies in rabbit; anti-VP6 serum antibody can be used as good diagnostic reagents for detection of PRV.
Figures



Similar articles
-
Development and Validation of Monoclonal Antibody-Based Antigen Capture ELISA for Detection of Group A Porcine Rotavirus.Viral Immunol. 2017 May;30(4):264-270. doi: 10.1089/vim.2016.0154. Epub 2017 Apr 17. Viral Immunol. 2017. PMID: 28414586
-
Development and evaluation of polyclonal antibodies based antigen capture ELISA for detection of porcine rotavirus.Anim Biotechnol. 2023 Nov;34(5):1807-1814. doi: 10.1080/10495398.2022.2052304. Epub 2022 May 20. Anim Biotechnol. 2023. PMID: 35593671
-
Development of a porcine epidemic diarrhea virus M protein-based ELISA for virus detection.Biotechnol Lett. 2011 Feb;33(2):215-20. doi: 10.1007/s10529-010-0420-8. Epub 2010 Sep 30. Biotechnol Lett. 2011. PMID: 20882317 Free PMC article.
-
Development of a rapid and sensitive latex agglutination-based method for detection of group A rotavirus.J Virol Methods. 2008 Mar;148(1-2):211-7. doi: 10.1016/j.jviromet.2007.11.013. Epub 2008 Jan 31. J Virol Methods. 2008. PMID: 18241934
-
Phylogenetic characterization of VP6 gene (inner capsid) of porcine rotavirus C collected in Japan.Infect Genet Evol. 2014 Aug;26:223-7. doi: 10.1016/j.meegid.2014.05.024. Epub 2014 Jun 11. Infect Genet Evol. 2014. PMID: 24929122
Cited by
-
Peptide-Recombinant VP6 Protein Based Enzyme Immunoassay for the Detection of Group A Rotaviruses in Multiple Host Species.PLoS One. 2016 Jul 8;11(7):e0159027. doi: 10.1371/journal.pone.0159027. eCollection 2016. PLoS One. 2016. PMID: 27391106 Free PMC article.
-
Triplex-Loop-Mediated Isothermal Amplification Combined with a Lateral Flow Immunoassay for the Simultaneous Detection of Three Pathogens of Porcine Viral Diarrhea Syndrome in Swine.Animals (Basel). 2023 Jun 7;13(12):1910. doi: 10.3390/ani13121910. Animals (Basel). 2023. PMID: 37370420 Free PMC article.
-
Design of ELISA-based diagnostic system for detection of enterohaemorrhagic Escherichia coli.Iran J Microbiol. 2025 Apr;17(2):278-286. doi: 10.18502/ijm.v17i2.18388. Iran J Microbiol. 2025. PMID: 40337686 Free PMC article.
-
Cholesterol of lipid rafts is a key determinant for entry and post-entry control of porcine rotavirus infection.BMC Vet Res. 2018 Feb 12;14(1):45. doi: 10.1186/s12917-018-1366-7. BMC Vet Res. 2018. PMID: 29433482 Free PMC article.
-
A novel enzyme-linked immunosorbent assay for detection of Escherichia coli O157:H7 using immunomagnetic and beacon gold nanoparticles.Gut Pathog. 2014 May 20;6:14. doi: 10.1186/1757-4749-6-14. eCollection 2014. Gut Pathog. 2014. PMID: 24864164 Free PMC article.
References
-
- Estes MK, Kapikian AZ. In: Fields virology. 5. Fields BN, Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editor. Vol. 10. Philadelphia, PA: Lippincott, Williams, and Wilkins; 2007. Rotaviruses and their replication, p.1917–1974.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

