Deciphering the Mechanisms of Developmental Disorders (DMDD): a new programme for phenotyping embryonic lethal mice
- PMID: 23519034
- PMCID: PMC3634640
- DOI: 10.1242/dmm.011957
Deciphering the Mechanisms of Developmental Disorders (DMDD): a new programme for phenotyping embryonic lethal mice
Abstract
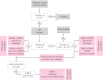
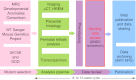
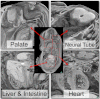
International efforts to test gene function in the mouse by the systematic knockout of each gene are creating many lines in which embryonic development is compromised. These homozygous lethal mutants represent a potential treasure trove for the biomedical community. Developmental biologists could exploit them in their studies of tissue differentiation and organogenesis; for clinical researchers they offer a powerful resource for investigating the origins of developmental diseases that affect newborns. Here, we outline a new programme of research in the UK aiming to kick-start research with embryonic lethal mouse lines. The 'Deciphering the Mechanisms of Developmental Disorders' (DMDD) programme has the ambitious goal of identifying all embryonic lethal knockout lines made in the UK over the next 5 years, and will use a combination of comprehensive imaging and transcriptomics to identify abnormalities in embryo structure and development. All data will be made freely available, enabling individual researchers to identify lines relevant to their research. The DMDD programme will coordinate its work with similar international efforts through the umbrella of the International Mouse Phenotyping Consortium [see accompanying Special Article (Adams et al., 2013)] and, together, these programmes will provide a novel database for embryonic development, linking gene identity with molecular profiles and morphology phenotypes.
Figures
References
-
- Adams D., Baldock R., Bhattacharya S., Copp A. J., Dickinson M., Greene N. D. E., Henkelman M., Justice M., Mohun T., Murray S. A., et al. (2013). Bloomsbury report on mouse embryo phenotyping: recommendations from the IMPC workshop on embryonic lethal screening. Dis. Model. Mech. 6, 571–579 - PMC - PubMed
-
- Aijö T., Lähdesmäki H. (2009). Learning gene regulatory networks from gene expression measurements using non-parametric molecular kinetics. Bioinformatics 25, 2937–2944 - PubMed
-
- Airik R., Kärner M., Karis A., Kärner J. (2005). Gene expression analysis of Gata3−/− mice by using cDNA microarray technology. Life Sci. 76, 2559–2568 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

